Themes: Difference between revisions
(Created page with "===Innate immune responses at the single cell level: Experiments and modeling=== Image:Themes-modeling_immune_responses.png|link=http://pmbm.ippt.gov.pl/images/d/df/Themes-...") |
mNo edit summary |
||
| Line 1: | Line 1: | ||
===Innate immune responses at the single cell level: Experiments and modeling=== | ===Innate immune responses at the single cell level: Experiments and modeling=== | ||
[[Image:Themes-modeling_immune_responses.png|link=http://pmbm.ippt.gov.pl/images/d/df/Themes-modeling_immune_responses.png|right|thumb| | [[Image:Themes-modeling_immune_responses.png|link=http://pmbm.ippt.gov.pl/images/d/df/Themes-modeling_immune_responses.png|right|thumb|360px|'''Example of pathogen recognition: LPS-induced signaling'''. LPS (lipopolysaccharide of the outer membrane of Gram-negative bacteria) is recognized by CD14 co-receptor, which transfers it to TLR4 resulting in its activation and binding of adaptor protein MyD88. Consequently, kinase TAK1 is activated and transmits signal to transcription factors p50-RelA (NF-κB) and AP-1 (early phase of ~30 min). CD14-induced endocytosis of CD14–TLR4–LPS complexes leads to TRIF-mediated activation of IRF3 and p50-RelA (late phase). Activation of transcription factors p50-RelA, IRF3 and AP1 leads to their nuclear translocation and synthesis of cytokines: TNFa, IL-1 and IFNβ that regulate via para/auto-crine signaling the second phase of innate immune responses. Transcriptional activity of p50-RelA and IRF3 is controlled by NF-κB-inducible proteins IkBα and A20 (negative feedbacks).]] | ||
Innate immunity forms the first line of defense, limiting spread of infection before the adaptive immune response is activated. In the first phase of the innate immune response, cells detect pathogens or their fragments with their membrane and cytoplasmic receptors. This leads to activation of the regulatory systems of the transcriptional factors of NF-κB, IRF3 and AP-1 families. These factors jointly regulate the activity of several hundred genes responsible for inducing inflammation, antiviral protection, proliferation, and apoptosis. In particular, they induce production and secretion of proinflammatory cytokines (among them IL-1, TNFα) as well as interferons-α and -β. These cytokines are mediators of the second phase of the cellular innate immune response in cells that did not encounter the pathogen. | Innate immunity forms the first line of defense, limiting spread of infection before the adaptive immune response is activated. In the first phase of the innate immune response, cells detect pathogens or their fragments with their membrane and cytoplasmic receptors. This leads to activation of the regulatory systems of the transcriptional factors of NF-κB, IRF3 and AP-1 families. These factors jointly regulate the activity of several hundred genes responsible for inducing inflammation, antiviral protection, proliferation, and apoptosis. In particular, they induce production and secretion of proinflammatory cytokines (among them IL-1, TNFα) as well as interferons-α and -β. These cytokines are mediators of the second phase of the cellular innate immune response in cells that did not encounter the pathogen. | ||
| Line 7: | Line 7: | ||
''Selected publications'': | ''Selected publications'': | ||
* <span class="nopmbm">Barua D</span>, <span class="nopmbm">Hlavacek WS</span>, <span class="pmbm">Lipniacki T</span>. '''A computational model for early events in B cell antigen receptor signaling: analysis of the roles of Lyn and Fyn''', ''J Immunol'' <u>189</u>:646-658 (2012) [http://www.ncbi.nlm.nih.gov/pubmed/22711887 PubMed] [http://dx.doi.org/10.4049/jimmunol.1102003 CrossRef] | [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3392547/pdf/nihms-378694.pdf PDF-ms] | * <span class="nopmbm">Barua D</span>, <span class="nopmbm">Hlavacek WS</span>, <span class="pmbm">Lipniacki T</span>. '''A computational model for early events in B cell antigen receptor signaling: analysis of the roles of Lyn and Fyn''', ''J Immunol'' <u>189</u>:646-658 (2012) [http://www.ncbi.nlm.nih.gov/pubmed/22711887 PubMed] [http://dx.doi.org/10.4049/jimmunol.1102003 CrossRef] | [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3392547/pdf/nihms-378694.pdf PDF-ms] | ||
* <span class="pmbm">Hat B</span>, <span class="pmbm">Kaźmierczak B</span>, <span class="pmbm">Lipniacki T</span>. '''B cell activation triggered by the formation of the small receptor cluster: a computational study''', ''PLoS Comput Biol'' <u>7</u>(10):e1000448 (2011) [http://www.ncbi.nlm.nih.gov/pubmed/21998572 PubMed] [http://dx.doi.org/10.1371/journal.pcbi.1002197 CrossRef] | [http://www.ploscompbiol.org/article/fetchObjectAttachment.action;jsessionid=0D25BED2B068B1B246EADA7099FD1A70?uri=info%3Adoi%2F10.1371%2Fjournal.pcbi.1002197&representation=PDF PDF] | * <span class="pmbm">Hat B</span>, <span class="pmbm">Kaźmierczak B</span>, <span class="pmbm">Lipniacki T</span>. '''B cell activation triggered by the formation of the small receptor cluster: a computational study''', ''PLoS Comput Biol'' <u>7</u>(10):e1000448 (2011) [http://www.ncbi.nlm.nih.gov/pubmed/21998572 PubMed] [http://dx.doi.org/10.1371/journal.pcbi.1002197 CrossRef] | [http://www.ploscompbiol.org/article/fetchObjectAttachment.action;jsessionid=0D25BED2B068B1B246EADA7099FD1A70?uri=info%3Adoi%2F10.1371%2Fjournal.pcbi.1002197&representation=PDF PDF] | ||
* <span class="nopmbm">Tay S</span>, <span class="nopmbm">Hughey J</span>, <span class="nopmbm">Lee T</span>, <span class="pmbm">Lipniacki T</span>, <span class="nopmbm">Covert M</span>, <span class="nopmbm">Quake S</span>. '''Single-cell NF-κB dynamics reveal digital activation and analogue information processing''', ''Nature'' <u>466</u>:267-271 (2010) [http://www.ncbi.nlm.nih.gov/pubmed/20581820 PubMed] [http://dx.doi.org/10.1038/nature09145 CrossRef] | [http://pmbm.ippt.pan.pl/publications/Tay-2010-Nature.pdf PDF] [http://pmbm.ippt.pan.pl/publications/supplementary/Tay-2010-Nature-SourceCode.zip Code] | * <span class="nopmbm">Tay S</span>, <span class="nopmbm">Hughey J</span>, <span class="nopmbm">Lee T</span>, <span class="pmbm">Lipniacki T</span>, <span class="nopmbm">Covert M</span>, <span class="nopmbm">Quake S</span>. '''Single-cell NF-κB dynamics reveal digital activation and analogue information processing''', ''Nature'' <u>466</u>:267-271 (2010) [http://www.ncbi.nlm.nih.gov/pubmed/20581820 PubMed] [http://dx.doi.org/10.1038/nature09145 CrossRef] | [http://pmbm.ippt.pan.pl/publications/Tay-2010-Nature.pdf PDF] [http://pmbm.ippt.pan.pl/publications/supplementary/Tay-2010-Nature-SourceCode.zip Code] | ||
<br /> | <br /> | ||
| Line 19: | Line 21: | ||
''Selected publications'': | ''Selected publications'': | ||
* <span class="nopmbm">Liepe J</span>, <span class="nopmbm">Filippi S</span>, <span class="pmbm">Komorowski M</span>, <span class="nopmbm">Stumpf MPH</span>. '''Maximising the information content of experiments in systems biology,''' ''PLOS Comput Biol'' <u>9</u>(1):e1002888 (2013) [http://www.ncbi.nlm.nih.gov/pubmed/23382663 PubMed] [http://10.1371/journal.pcbi.1002888 CrossRef] | * <span class="nopmbm">Liepe J</span>, <span class="nopmbm">Filippi S</span>, <span class="pmbm">Komorowski M</span>, <span class="nopmbm">Stumpf MPH</span>. '''Maximising the information content of experiments in systems biology,''' ''PLOS Comput Biol'' <u>9</u>(1):e1002888 (2013) [http://www.ncbi.nlm.nih.gov/pubmed/23382663 PubMed] [http://10.1371/journal.pcbi.1002888 CrossRef] | ||
* <span class="pmbm">Komorowski M</span>, <span class="nopmbm">Costa MJ</span>, <span class="nopmbm">Rand DA</span>, <span class="nopmbm">Stumpf MPH</span>. '''Sensitivity, robustness, and identifiability in stochastic chemical kinetics models,''' ''Proc Natl Acad Sci USA'' <u>108</u>(21):8645-50 (2011) [http://www.ncbi.nlm.nih.gov/pubmed/21551095 PubMed] [http://dx.doi.org/10.1073/pnas.1015814108 CrossRef] | * <span class="pmbm">Komorowski M</span>, <span class="nopmbm">Costa MJ</span>, <span class="nopmbm">Rand DA</span>, <span class="nopmbm">Stumpf MPH</span>. '''Sensitivity, robustness, and identifiability in stochastic chemical kinetics models,''' ''Proc Natl Acad Sci USA'' <u>108</u>(21):8645-50 (2011) [http://www.ncbi.nlm.nih.gov/pubmed/21551095 PubMed] [http://dx.doi.org/10.1073/pnas.1015814108 CrossRef] | ||
* <span class="pmbm">Komorowski M</span>, <span class="nopmbm">Finkenstädt B</span>, <span class="nopmbm">Harper CV</span>, <span class="nopmbm">Rand DA</span>. '''Bayesian inference of biochemical kinetic parameters using the linear noise approximation,''' ''BMC Bioinformatics'' <u>10</u>:343 (2009) [http://www.ncbi.nlm.nih.gov/pubmed/19840370 PubMed] [http://dx.doi.org/10.1186/1471-2105-10-343 CrossRef] | * <span class="pmbm">Komorowski M</span>, <span class="nopmbm">Finkenstädt B</span>, <span class="nopmbm">Harper CV</span>, <span class="nopmbm">Rand DA</span>. '''Bayesian inference of biochemical kinetic parameters using the linear noise approximation,''' ''BMC Bioinformatics'' <u>10</u>:343 (2009) [http://www.ncbi.nlm.nih.gov/pubmed/19840370 PubMed] [http://dx.doi.org/10.1186/1471-2105-10-343 CrossRef] | ||
<br /> | <br /> | ||
| Line 30: | Line 34: | ||
''Selected publications'': | ''Selected publications'': | ||
* <span class="nopmbm">Węgrzyn J</span>, <span class="nopmbm">Samborski A</span>, <span class="nopmbm">Reissig L</span>, <span class="pmbm">Korczyk PM</span>, <span class="pmbm">Błoński S</span>, <span class="nopmbm">Garstecki P</span>. '''Microfluidic architectures for efficient generation of chemistry gradations in droplets''', ''Microfluid Nanofluid'' <u>14</u>(1-2):235–245 (2013, e-pub 2012) [http://dx.doi.org/10.1007/s10404-012-1042-3 CrossRef] | [http://pmbm.ippt.pan.pl/publications/Wegrzyn-2013-MicrofluidNanofluid.pdf PDF] | * <span class="nopmbm">Węgrzyn J</span>, <span class="nopmbm">Samborski A</span>, <span class="nopmbm">Reissig L</span>, <span class="pmbm">Korczyk PM</span>, <span class="pmbm">Błoński S</span>, <span class="nopmbm">Garstecki P</span>. '''Microfluidic architectures for efficient generation of chemistry gradations in droplets''', ''Microfluid Nanofluid'' <u>14</u>(1-2):235–245 (2013, e-pub 2012) [http://dx.doi.org/10.1007/s10404-012-1042-3 CrossRef] | [http://pmbm.ippt.pan.pl/publications/Wegrzyn-2013-MicrofluidNanofluid.pdf PDF] | ||
* <span class="nopmbm">Jakiela S</span>, <span class="pmbm">Korczyk PM</span>, <span class="nopmbm">Makulska S</span>, <span class="nopmbm">Cybulski O</span>, <span class="nopmbm">Garstecki P</span>. '''Discontinuous transition in a laminar fluid flow: A change of flow topolgy inside a droplet moving in a micron-size channel''', ''Phys Rev Lett'' <u>108</u>(13):134501 (2012) [http://www.ncbi.nlm.nih.gov/pubmed/22540703 PubMed] [http://dx.doi.org/10.1103/PhysRevLett.108.134501 CrossRef] | * <span class="nopmbm">Jakiela S</span>, <span class="pmbm">Korczyk PM</span>, <span class="nopmbm">Makulska S</span>, <span class="nopmbm">Cybulski O</span>, <span class="nopmbm">Garstecki P</span>. '''Discontinuous transition in a laminar fluid flow: A change of flow topolgy inside a droplet moving in a micron-size channel''', ''Phys Rev Lett'' <u>108</u>(13):134501 (2012) [http://www.ncbi.nlm.nih.gov/pubmed/22540703 PubMed] [http://dx.doi.org/10.1103/PhysRevLett.108.134501 CrossRef] | ||
* <span class="pmbm">Korczyk PM</span>, <span class="nopmbm">Cybulski O</span>, <span class="nopmbm">Makulska S</span>, <span class="nopmbm">Garstecki P</span>. '''Effects of unsteadiness of the rates of flow on the dynamics of formation of droplets in microfluidic systems''', ''Lab Chip'' <u>11</u>:173-175 (2011) [http://www.ncbi.nlm.nih.gov/pubmed/20949204 PubMed] [http://dx.doi.org/10.1039/C0LC00088D CrossRef] | * <span class="pmbm">Korczyk PM</span>, <span class="nopmbm">Cybulski O</span>, <span class="nopmbm">Makulska S</span>, <span class="nopmbm">Garstecki P</span>. '''Effects of unsteadiness of the rates of flow on the dynamics of formation of droplets in microfluidic systems''', ''Lab Chip'' <u>11</u>:173-175 (2011) [http://www.ncbi.nlm.nih.gov/pubmed/20949204 PubMed] [http://dx.doi.org/10.1039/C0LC00088D CrossRef] | ||
<br /> | <br /> | ||
| Line 44: | Line 50: | ||
''Selected publications'': | ''Selected publications'': | ||
* <span class="pmbm">Kaźmierczak B</span>, <span class="pmbm">Peradzyński Z</span>. '''Calcium waves with fast buffers and mechanical effects''', ''J Math Biol'' <u>62</u>:1-38 (2011, online 2010) [http://www.ncbi.nlm.nih.gov/pubmed/20098991 PubMed] [http://dx.doi.org/10.1007/s00285-009-0323-2 CrossRef] | * <span class="pmbm">Kaźmierczak B</span>, <span class="pmbm">Peradzyński Z</span>. '''Calcium waves with fast buffers and mechanical effects''', ''J Math Biol'' <u>62</u>:1-38 (2011, online 2010) [http://www.ncbi.nlm.nih.gov/pubmed/20098991 PubMed] [http://dx.doi.org/10.1007/s00285-009-0323-2 CrossRef] | ||
* <span class="pmbm">Kaźmierczak B</span>, <span class="nopmbm">Volpert V</span>. '''Calcium waves in systems with immobile buffers as a limit of waves for systems with non zero diffusion''', ''Nonlinearity'' <u>21</u>:71 (2008) [http://dx.doi.org/10.1088/0951-7715/21/1/004 CrossRef] | * <span class="pmbm">Kaźmierczak B</span>, <span class="nopmbm">Volpert V</span>. '''Calcium waves in systems with immobile buffers as a limit of waves for systems with non zero diffusion''', ''Nonlinearity'' <u>21</u>:71 (2008) [http://dx.doi.org/10.1088/0951-7715/21/1/004 CrossRef] | ||
* <span class="pmbm">Kaźmierczak B</span>, <span class="nopmbm">Volpert V</span>. '''Travelling calcium waves in systems with non diffusing buffers''', ''Math Mod Meth Appl Sci'' <u>18</u>(6):883-912 (2008) [http://dx.doi.org/10.1142/S0218202508002899 CrossRef] | * <span class="pmbm">Kaźmierczak B</span>, <span class="nopmbm">Volpert V</span>. '''Travelling calcium waves in systems with non diffusing buffers''', ''Math Mod Meth Appl Sci'' <u>18</u>(6):883-912 (2008) [http://dx.doi.org/10.1142/S0218202508002899 CrossRef] | ||
<br /> | <br /> | ||
| Line 55: | Line 63: | ||
''Selected publications'': | ''Selected publications'': | ||
* <span class="pmbm">Kochańczyk M</span>, <span class="pmbm">Jaruszewicz J</span>, <span class="pmbm">Lipniacki T</span>. '''Stochastic transitions in a bistable reaction system on the membrane''', ''J R Soc Interface'' <u>10</u>(84):20130151 (2013) [http://www.ncbi.nlm.nih.gov/pubmed/23635492 PubMed] [http://dx.doi.org/10.1098/rsif.2013.0151 CrossRef] | [http://rsif.royalsocietypublishing.org/content/10/84/20130151.full.pdf PDF] [http://pmbm.ippt.pan.pl/publications/supplementary/Kochanczyk-2013-JRSocInterface-Supp.pdf Supp-PDF] [http://pmbm.ippt.pan.pl/publications/supplementary/Kochanczyk-2013-JRSocInterface-Movies.zip Supp-movies] | * <span class="pmbm">Kochańczyk M</span>, <span class="pmbm">Jaruszewicz J</span>, <span class="pmbm">Lipniacki T</span>. '''Stochastic transitions in a bistable reaction system on the membrane''', ''J R Soc Interface'' <u>10</u>(84):20130151 (2013) [http://www.ncbi.nlm.nih.gov/pubmed/23635492 PubMed] [http://dx.doi.org/10.1098/rsif.2013.0151 CrossRef] | [http://rsif.royalsocietypublishing.org/content/10/84/20130151.full.pdf PDF] [http://pmbm.ippt.pan.pl/publications/supplementary/Kochanczyk-2013-JRSocInterface-Supp.pdf Supp-PDF] [http://pmbm.ippt.pan.pl/publications/supplementary/Kochanczyk-2013-JRSocInterface-Movies.zip Supp-movies] | ||
* <span class="pmbm">Żuk PJ</span>, <span class="pmbm">Kochańczyk M</span>, <span class="pmbm">Jaruszewicz J</span>, <span class="nopmbm">Bednorz W</span>, <span class="pmbm">Lipniacki T</span>. '''Dynamics of a stochastic spatially extended system predicted by comparing deterministic and stochastic attractors of the corresponding birth-death process''', ''Phys Biol'' <u>5</u>(9):055002 (2012) [http://www.ncbi.nlm.nih.gov/pubmed/23011381 PubMed] [http://dx.doi.org/10.1088/1478-3975/9/5/055002 CrossRef] | [http://pmbm.ippt.pan.pl/publications/Zuk-2012-PhysBiol-MS.pdf PDF-ms] | * <span class="pmbm">Żuk PJ</span>, <span class="pmbm">Kochańczyk M</span>, <span class="pmbm">Jaruszewicz J</span>, <span class="nopmbm">Bednorz W</span>, <span class="pmbm">Lipniacki T</span>. '''Dynamics of a stochastic spatially extended system predicted by comparing deterministic and stochastic attractors of the corresponding birth-death process''', ''Phys Biol'' <u>5</u>(9):055002 (2012) [http://www.ncbi.nlm.nih.gov/pubmed/23011381 PubMed] [http://dx.doi.org/10.1088/1478-3975/9/5/055002 CrossRef] | [http://pmbm.ippt.pan.pl/publications/Zuk-2012-PhysBiol-MS.pdf PDF-ms] | ||
* <span class="pmbm">Szymańska P</span>, <span class="pmbm">Kochańczyk M</span>, <span class="nopmbm">Miększ J</span>, <span class="pmbm">Lipniacki T</span>. '''Effective reaction rates in diffusion-limited phosphorylation–dephosphorylation cycles''', ''Phys Rev E'' <u>91</u>:022702 (2015) [http://dx.doi.org/10.1103/PhysRevE.91.022702 CrossRef] | [http://pmbm.ippt.pan.pl/publications/Szymanska-2015-PhysRevE-MS.pdf PDF-ms] | * <span class="pmbm">Szymańska P</span>, <span class="pmbm">Kochańczyk M</span>, <span class="nopmbm">Miększ J</span>, <span class="pmbm">Lipniacki T</span>. '''Effective reaction rates in diffusion-limited phosphorylation–dephosphorylation cycles''', ''Phys Rev E'' <u>91</u>:022702 (2015) [http://dx.doi.org/10.1103/PhysRevE.91.022702 CrossRef] | [http://pmbm.ippt.pan.pl/publications/Szymanska-2015-PhysRevE-MS.pdf PDF-ms] | ||
| Line 68: | Line 78: | ||
* <span class="nopmbm">Klosowski T</span>, <span class="pmbm">Kowalczyk T</span>, <span class="nopmbm">Nowacki M</span>, <span class="nopmbm">Drewa T</span>. '''Tissue engineering and ureter regeneration, is it possible?''' ''Int J Artif Organs'' <u>36</u>(6):392-405 (2013) [http://www.ncbi.nlm.nih.gov/pubmed/23645581 PubMed] [http://dx.doi.org/10.5301/ijao.5000130 CrossRef] | * <span class="nopmbm">Klosowski T</span>, <span class="pmbm">Kowalczyk T</span>, <span class="nopmbm">Nowacki M</span>, <span class="nopmbm">Drewa T</span>. '''Tissue engineering and ureter regeneration, is it possible?''' ''Int J Artif Organs'' <u>36</u>(6):392-405 (2013) [http://www.ncbi.nlm.nih.gov/pubmed/23645581 PubMed] [http://dx.doi.org/10.5301/ijao.5000130 CrossRef] | ||
* <span class="nopmbm">Bretcanu O</span>, <span class="nopmbm">Misra SK</span>, <span class="nopmbm">Yunos DM</span>, <span class="nopmbm">Boccaccini AR</span>, <span class="nopmbm">Roy I</span>, <span class="pmbm">Kowalczyk T</span>, <span class="pmbm">Błoński S</span>, <span class="pmbm">Kowalewski TA</span>. '''Electrospun nanofibrous biodegradable polyester coatings on Bioglass-based glass-ceramics for tissue engineering''', ''Mater Chem Phys'' <u>118</u>:420-426 (2009) [http://dx.doi.org/10.1016/j.matchemphys.2009.08.011 CrossRef] | * <span class="nopmbm">Bretcanu O</span>, <span class="nopmbm">Misra SK</span>, <span class="nopmbm">Yunos DM</span>, <span class="nopmbm">Boccaccini AR</span>, <span class="nopmbm">Roy I</span>, <span class="pmbm">Kowalczyk T</span>, <span class="pmbm">Błoński S</span>, <span class="pmbm">Kowalewski TA</span>. '''Electrospun nanofibrous biodegradable polyester coatings on Bioglass-based glass-ceramics for tissue engineering''', ''Mater Chem Phys'' <u>118</u>:420-426 (2009) [http://dx.doi.org/10.1016/j.matchemphys.2009.08.011 CrossRef] | ||
* <span class="pmbm">Kowalczyk T</span>, <span class="nopmbm">Nowicka A</span>, <span class="nopmbm">Elbaum D</span>, <span class="pmbm">Kowalewski TA</span>. '''Electrospinning of bovine serum albumin. Optimization and the use for production of biosensors''', ''Biomacromolecules'' <u>9</u>:2087-2090 (2008) [http://www.ncbi.nlm.nih.gov/pubmed/18576601 PubMed] [http://dx.doi.org/10.1021/bm800421s CrossRef] | * <span class="pmbm">Kowalczyk T</span>, <span class="nopmbm">Nowicka A</span>, <span class="nopmbm">Elbaum D</span>, <span class="pmbm">Kowalewski TA</span>. '''Electrospinning of bovine serum albumin. Optimization and the use for production of biosensors''', ''Biomacromolecules'' <u>9</u>:2087-2090 (2008) [http://www.ncbi.nlm.nih.gov/pubmed/18576601 PubMed] [http://dx.doi.org/10.1021/bm800421s CrossRef] | ||
| Line 81: | Line 93: | ||
''Selected publications'': | ''Selected publications'': | ||
* <span class="pmbm">Szczepański J</span>, <span class="nopmbm">Arnold M</span>, <span class="nopmbm">Wajnryb E</span>, <span class="nopmbm">Amigó JM</span>, <span class="nopmbm">Sanchez-Vives MV</span>. '''Mutual information and redundancy in spontaneous communication between cortical neurons''', ''Biol Cybern'' <u>104</u>:161-174 (2011) [http://www.ncbi.nlm.nih.gov/pubmed/21340601 PubMed] [http://10.1007/s00422-011-0425-y CrossRef] | * <span class="pmbm">Szczepański J</span>, <span class="nopmbm">Arnold M</span>, <span class="nopmbm">Wajnryb E</span>, <span class="nopmbm">Amigó JM</span>, <span class="nopmbm">Sanchez-Vives MV</span>. '''Mutual information and redundancy in spontaneous communication between cortical neurons''', ''Biol Cybern'' <u>104</u>:161-174 (2011) [http://www.ncbi.nlm.nih.gov/pubmed/21340601 PubMed] [http://10.1007/s00422-011-0425-y CrossRef] | ||
* <span class="nopmbm">Paprocki B</span>, <span class="pmbm">Szczepański J</span>. '''Efficiency of neural transmission as a function of synaptic noise, threshold, and source characteristics''', ''Biosystems'' <u>105</u>(1):62-72 (2011) [http://www.ncbi.nlm.nih.gov/pubmed/21439348 PubMed] [http://dx.doi.org/10.1016/j.biosystems.2011.03.005 CrossRef] | * <span class="nopmbm">Paprocki B</span>, <span class="pmbm">Szczepański J</span>. '''Efficiency of neural transmission as a function of synaptic noise, threshold, and source characteristics''', ''Biosystems'' <u>105</u>(1):62-72 (2011) [http://www.ncbi.nlm.nih.gov/pubmed/21439348 PubMed] [http://dx.doi.org/10.1016/j.biosystems.2011.03.005 CrossRef] | ||
* <span class="nopmbm">Nagarajan R</span>, <span class="pmbm">Szczepański J</span>, <span class="nopmbm">Wajnryb E</span>. '''Interpreting non-random signatures in biomedical signals using Lempel-Ziv complexity''', ''Physica D'' <u>237</u>(3):359-364 (2008) [http://dx.doi.org/10.1016/j.physd.2007.09.007 CrossRef] | * <span class="nopmbm">Nagarajan R</span>, <span class="pmbm">Szczepański J</span>, <span class="nopmbm">Wajnryb E</span>. '''Interpreting non-random signatures in biomedical signals using Lempel-Ziv complexity''', ''Physica D'' <u>237</u>(3):359-364 (2008) [http://dx.doi.org/10.1016/j.physd.2007.09.007 CrossRef] | ||
--> | --> | ||
<br /> | <br /> | ||
<br /> | <br /> | ||
Revision as of 15:19, 16 June 2017
Innate immune responses at the single cell level: Experiments and modeling
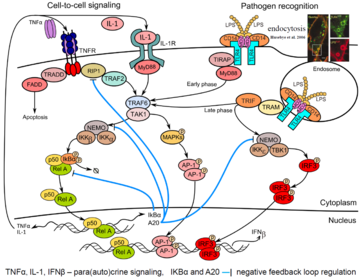
Innate immunity forms the first line of defense, limiting spread of infection before the adaptive immune response is activated. In the first phase of the innate immune response, cells detect pathogens or their fragments with their membrane and cytoplasmic receptors. This leads to activation of the regulatory systems of the transcriptional factors of NF-κB, IRF3 and AP-1 families. These factors jointly regulate the activity of several hundred genes responsible for inducing inflammation, antiviral protection, proliferation, and apoptosis. In particular, they induce production and secretion of proinflammatory cytokines (among them IL-1, TNFα) as well as interferons-α and -β. These cytokines are mediators of the second phase of the cellular innate immune response in cells that did not encounter the pathogen.
Selected publications:
- Barua D, Hlavacek WS, Lipniacki T. A computational model for early events in B cell antigen receptor signaling: analysis of the roles of Lyn and Fyn, J Immunol 189:646-658 (2012) PubMed CrossRef | PDF-ms
- Hat B, Kaźmierczak B, Lipniacki T. B cell activation triggered by the formation of the small receptor cluster: a computational study, PLoS Comput Biol 7(10):e1000448 (2011) PubMed CrossRef | PDF
- Tay S, Hughey J, Lee T, Lipniacki T, Covert M, Quake S. Single-cell NF-κB dynamics reveal digital activation and analogue information processing, Nature 466:267-271 (2010) PubMed CrossRef | PDF Code
Statistical methods to investigate biochemical dynamics
Reverse-engineering of biochemical networks is largely dependent on the quality of available data. We are interested in the development of mathematical tools to help wet experimentation in obtaining more insight about the latent mechanisms that generate experimentally observed dynamics. Specifically, we incorporate available knowledge to find experimental conditions which have a potential to reveal missing information about the studied systems. We use a variety of statistical methods ranging from bayesian statistics, through model selection, to information geometry.
Selected publications:
- Liepe J, Filippi S, Komorowski M, Stumpf MPH. Maximising the information content of experiments in systems biology, PLOS Comput Biol 9(1):e1002888 (2013) PubMed CrossRef
- Komorowski M, Costa MJ, Rand DA, Stumpf MPH. Sensitivity, robustness, and identifiability in stochastic chemical kinetics models, Proc Natl Acad Sci USA 108(21):8645-50 (2011) PubMed CrossRef
- Komorowski M, Finkenstädt B, Harper CV, Rand DA. Bayesian inference of biochemical kinetic parameters using the linear noise approximation, BMC Bioinformatics 10:343 (2009) PubMed CrossRef
Microfluidics for biomedical research and applications
Microfluidics have become an important tool in research and variety of industrial applications. Microfluidic devices enable precise operations on small volumes of liquids which can be obtain by the use of “smart” geometries or by the control of external devices. On the field of biological researches microfluidic devices can mimic physiological conditions better than other in vitro systems, and can be used for long term cell culture. They are controlled with high precision by external devices such pumps, valves and detectors, and can be easily automated. These features enable high throughput screening of the effects of different mixtures of reagents on the behaviour of cells, rendering microfluidics a powerful tool in research in cell biology and medicine. Microfluidics enables also the use of small droplets as separated chemical reactors or incubators. This approach which will be developed in the future enables to close the population of cells into small separated volume in which concentration of any reagent can be precisely adjusted.
Selected publications:
- Węgrzyn J, Samborski A, Reissig L, Korczyk PM, Błoński S, Garstecki P. Microfluidic architectures for efficient generation of chemistry gradations in droplets, Microfluid Nanofluid 14(1-2):235–245 (2013, e-pub 2012) CrossRef | PDF
- Jakiela S, Korczyk PM, Makulska S, Cybulski O, Garstecki P. Discontinuous transition in a laminar fluid flow: A change of flow topolgy inside a droplet moving in a micron-size channel, Phys Rev Lett 108(13):134501 (2012) PubMed CrossRef
- Korczyk PM, Cybulski O, Makulska S, Garstecki P. Effects of unsteadiness of the rates of flow on the dynamics of formation of droplets in microfluidic systems, Lab Chip 11:173-175 (2011) PubMed CrossRef
Modeling of calcium signaling pathways
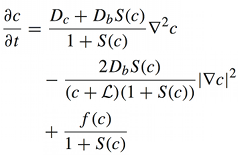
Calcium is one of the most important signaling messengers. Fundamental cellular processes are regulated by the influx of calcium from the extracellular matrix or its transport between intracellular compartments, e.g. cytosol ↔ mitochondria, mitochondria ↔ ER.
Selected publications:
- Kaźmierczak B, Peradzyński Z. Calcium waves with fast buffers and mechanical effects, J Math Biol 62:1-38 (2011, online 2010) PubMed CrossRef
- Kaźmierczak B, Volpert V. Calcium waves in systems with immobile buffers as a limit of waves for systems with non zero diffusion, Nonlinearity 21:71 (2008) CrossRef
- Kaźmierczak B, Volpert V. Travelling calcium waves in systems with non diffusing buffers, Math Mod Meth Appl Sci 18(6):883-912 (2008) CrossRef
Kinetic Monte Carlo simulations of biochemical reactions on the plasma membrane
Early events in cellular communication and signaling engage proteins located in the plasma membrane. Slow diffusion therein restricts the number of interacting molecules, enhancing molecular noise, and renders the membrane a spatially organized stochastic reactor. Therefore, numerical simulation of signaling on the membrane requires methods which can account for both spatial resolution and stochastic effects, such as the kinetic Monte Carlo on the lattice.
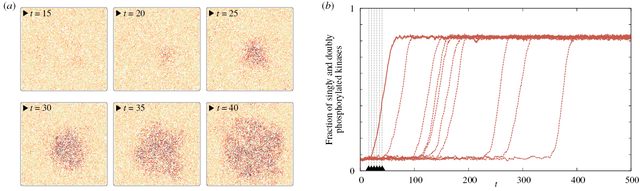
Selected publications:
- Kochańczyk M, Jaruszewicz J, Lipniacki T. Stochastic transitions in a bistable reaction system on the membrane, J R Soc Interface 10(84):20130151 (2013) PubMed CrossRef | PDF Supp-PDF Supp-movies
- Żuk PJ, Kochańczyk M, Jaruszewicz J, Bednorz W, Lipniacki T. Dynamics of a stochastic spatially extended system predicted by comparing deterministic and stochastic attractors of the corresponding birth-death process, Phys Biol 5(9):055002 (2012) PubMed CrossRef | PDF-ms
- Szymańska P, Kochańczyk M, Miększ J, Lipniacki T. Effective reaction rates in diffusion-limited phosphorylation–dephosphorylation cycles, Phys Rev E 91:022702 (2015) CrossRef | PDF-ms