Grants: Difference between revisions
mNo edit summary |
mNo edit summary |
||
| (123 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
<ul class="nav nav-tabs"> | <ul class="nav nav-tabs mb-2" role="tablist"> | ||
<li class="nav-item"><btn class="nav-link active" data-toggle="tab" role="tab" aria-controls="CurrentGrants" aria-selected="true">#CurrentGrants|===Current===</btn></li> | |||
<li class="nav-item"><btn class="nav-link" data-toggle="tab" role="tab" aria-controls="PastGrants" aria-selected="false">#PastGrants|===Past===</btn></li> | |||
<li class="nav-item"><btn class="nav-link" data-toggle="tab" role="tab" aria-controls="TEAM" aria-selected="false">#TEAM|===FNP TEAM===</btn></li> | |||
<li class="nav-item"><btn lass="nav-link" data-toggle="tab" role="tab" aria-controls="GRIEG" aria-selected="false">#GRIEG|===GRIEG NK cells===</btn></li> | |||
</ul> | </ul> | ||
<div class="tab-content"> | <div class="tab-content"> | ||
<div id="CurrentGrants" class="tab-pane fade show active" role="tabpanel"> | |||
* '' | *''Combinatorial targeting of non-genetic sources of cell-to-cell heterogeneity to overcome fractional killing of cancer cells''.<br/>National Science Centre (NCN): Opus, 2022–2025.<br/>PI: Jarosław Walczak | ||
* '' | *''Cell-to-cell heterogeneity of cross-wired cytokine signaling: filling the gaps between molecular origins and translational implications''.<br/>National Science Centre (NCN): Opus, 2021–2025.<br/>PI: Michał Komorowski | ||
* '' | *''Interaction of natural killer cells with an infected cancer cell population: microfluidics-based experiments and single-cell-level mathematical modeling''.<br/>National Science Centre (NCN): '''Grieg''' (Norwegian Financial Mechanism), 2020–2023.<br/>PI: Tomasz Lipniacki | ||
* '' | *''Influence of oncogenic mutations on information transmission in the MAPK signaling pathway''.<br/>National Science Centre (NCN): Opus, 2020–2024.<br/>PI: Tomasz Lipniacki | ||
* '' | *''Sprayable multifunctional nanofibrous hydrogels for polytherapeutic wound healing treatments''.<br/>National Centre for Research and Development (NCBiR): 2nd Polish–Chinese/Chinese–Polish Joint Research Call, 2021–2024.<br/>PI: Filippo Pierini | ||
</div> <!-- CurrentGrants --> | |||
<div id="PastGrants" class="tab-pane fade" role="tabpanel"> | |||
* '' | *''Electrospun conducting hydrogel nanomaterials for neural tissue engineering''.<br/>Foundation for Polish Science ('''FNP'''): '''First Team''', 2019–2022.<br/>PI: Filippo Pierini | ||
* '' | *''Coordination of innate immune response in infected cell population: experiment and mathematical modeling''.<br/>National Science Centre (NCN): Opus 15, 2019–2022.<br/>PI: Tomasz Lipniacki | ||
* '' | *''Deciphering biochemical signalling to inform more efficient therapeutic strategies''.<br/>Foundation for Polish Science ('''FNP'''): '''First Team''', 2018–2021<br/>PI: Michał Komorowski | ||
* '' | *''Development of novel methods for measuring viscosity and interfacial tension of liquids using microfluidic devices''.<br/>National Science Centre (NCN): Preludium 15, 2019–2022.<br/>PI: Damian Zaremba | ||
* '' | *''Exlaining intercellular heterogeneity of the response to IFNγ''.<br/>National Science Centre (NCN): Preludium 17, 2020–2022.<br/>PI: Piotr Topolewski | ||
* '' | *''Injectible cell scaffolds in tissue engineering''.<br/>National Centre for Research and Development (NCBiR): Lider 9, 2019–2022.<br/>PI: Paweł Nakielski | ||
*''Self-organization and nano-mechanical characterization of cellulose microfibers''.<br/>National Science Centre (NCN): Sonata 13, 2018–2021.<br/>PI: Adolfo Poma Bernaola | |||
* '' | |||
* '' | *''Digital operations on droplets embedded into smart microfluidic architectures for applications in medical diagnostics and biological research''.<br/>Foundation for Polish Science ('''FNP'''): '''First Team''', 2018–2020.<br/>PI: Piotr Korczyk | ||
* '' | *''Design, construction and optimization of a combined Atomic Force Microscope and Optical Tweezers instrument for single molecules and nanomaterials characterization''.<br/>National Centre for Research and Development (NCBiR): Lider 7, 2017–2020.<br/>PI: Filippo Pierini | ||
* '' | *''The effect of shear flow on fibrin clot structure and fibrin-platelets interactions''.<br/>Foundation for Polish Science ('''FNP'''): '''Homing''', 2017–2020.<br/>PI: Izabela Piechocka | ||
* '' | *''Non-standard parabolic problems in description of biological processes''.<br/>National Science Centre (NCN): OPUS 11, 2017–2020.<br/>PI: Bogdan Kaźmierczak | ||
* '' | *''Innate immune response to RSV infection, role of non-structural proteins. Experiments and modeling of signal transduction and among cells communication.''<br/>National Science Centre (NCN): Harmonia 6, 2015–2019.<br/>PI: Tomasz Lipniacki | ||
* '' | *''Stimuli-responsive chiral nematic liquid crystal hydrogel implants by electrospinning technique''.<br/>National Science Centre (NCN): Sonata 10, 2016–2019.<br/>PI: Filippo Pierini | ||
* '' | *''Investigation of blood clotting mechanism in the contact with nanofibers''.<br/>National Science Centre (NCN): Sonata 10, 2016–2019.<br/>PI: Paweł Nakielski | ||
* '' | *''Multi-scale dynamics of signal transduction: dissecting the MAPK pathway''.<br/>Wiener Wissenschafts-, Forschungs- und Technologiefonds (WWTF Austria) grant "Mathematics and...", 2015–2018.<br/>Co-PI: Tomasz Lipniacki | ||
* ''Efficient methods for generation of secure parameters of public key algorithms '' (''Opracowanie wydajnych metod generowania bezpiecznych parametrów algorytmów klucza publicznego'').<br />National Science | *''Dynamics of STAT1, STAT3, and STAT5 phosphorylation in breast cancer cell lines: a systematic theoretical and experimental analysis''.<br/>National Science Centre (NCN): Opus 9, 2016–2019.<br/>PI: Michał Komorowski | ||
*''Detecting signalling aberrations in cancer cells''.<br/>National Science Centre (NCN): Preludium 12, 2017–2019.<br/>PI: Karol Nienałtowski | |||
*''Analysis of hydrodynamic interactions between single molecules using optical tweezers''.<br/>National Science Centre (NCN): Preludium 10, 2016–2019.<br/>PI: Krzysztof Zembrzycki | |||
*''Mechanisms controlling cell fate decisions in response to stress. Experimental and theoretical analysis.''<br/>National Science Centre (NCN): Opus 7, 2015–2018.<br/>PI: Tomasz Lipniacki. | |||
*''The study of hydrodynamic interactions of droplets in complex microfluidic structures. The analysis of algorithms encoded in the architecture of microchannels enabling dosing of the reactants with arbitrary precision''.<br/>National Science Centre (NCN): Sonata Bis 4, 2015–2018.<br/>PI: Piotr Korczyk | |||
*European Molecular Biology Organisation ('''EMBO'''): '''Installation Grant''', 2013–2016/2018.<br/>PI: Michał Komorowski | |||
*''Hydrogel noanofilaments for biomedical application''.<br/>National Science Centre (NCN): Preludium 9, 2016–2018.<br/>PI: Sylwia Pawłowska | |||
*''Innate immune signalling: optimal microfluidic protocols, prediction and control''.<br/>Marie Curie Actions: Career Integration Grant, 2013–2017.<br/>PI: Michał Komorowski. | |||
*''Stochastic phenotype switching in growing and dividing bacteria.''<br/>National Science Centre (NCN): Preludium 7, 2015–2017.<br/>PI: Joanna Jaruszewicz–Błońska | |||
*''Regulation of the RAF/MEK/ERK cascade involving RAF isoforms''.<br/>National Science Centre (NCN): Preludium 6, 2014–2017.<br/>PI: Paweł Kocieniewski | |||
*''Dynamics of micro and nano objects suspended in fluids''.<br/>National Science Centre (NCN): OPUS 2, 2012–2016.<br/>PI: Tomasz Kowalewski | |||
*''Formation of spatial heterogeneities on the plasma membrane and their impact on cell signaling''.<br/>National Science Centre (NCN): Preludium 5, 2014–2016.<br/>PI: Marek Kochańczyk | |||
*''Inter- and intracellular signaling in innate immune response: experimental and mathematical analysis''.<br/>National Science Centre (NCN): OPUS 2, 2012–2015.<br/>PI: Tomasz Lipniacki | |||
*''Estimation of the kinetic rates in the NF-κΒ signalling system''.<br/>Polish Ministry of Science and Higher Education: Iuventus Plus, 2013–2015.<br/>PI: Michał Komorowski | |||
*''Mechanistic aspects and spatial effects in cell signaling''.<br/>Foundation for Polish Science '''(FNP): TEAM''', 2009–2013.<br/>PI: Tomasz Lipniacki | |||
*''Multiparameter and information theoretic models of biochemical signal transduction – modeling and inference''.<br/>Foundation for Polish Science (FNP): HOMING Plus, 2011–2013.<br/>PI: Michał Komorowski | |||
*''Application of electrospun nanofibrous mats as active wound dressing for prevention of post-traumatic brain damage'' (''Zastosowanie elektroprzędzonych nanowłókien jako opatrunków aktywnych w zapobieganiu pourazowym zmianom w tkance mózgowej''). <br/>National Center for Research and Development (NCBiR), 2010–2013.<br/>PI: Tomasz Kowalewski | |||
*''Traveling waves in cylindrical domains'' (''Fale biegnące w obszarach cylindrycznych''). National Science Centre (NCN), 2010–2012.<br/>PI: Bogdan Kaźmierczak | |||
*''Application of information theory-based computational methods for the analysis of signal transduction efficiency in neuronal networks'' (''Zastosowanie metod komputerowych bazujących na teorii informacji do analizy efektywności transmisji sygnałów w sieciach neuronowych'').<br/>National Science Centre (NCN), 2011–2012.<br/>PI: Janusz Szczepański | |||
*''Efficient methods for generation of secure parameters of public key algorithms'' (''Opracowanie wydajnych metod generowania bezpiecznych parametrów algorytmów klucza publicznego'').<br/>National Science Centre (NCN): grant for a Ph.D. student, 2011–2012.<br/>PI: Janusz Szczepański | |||
</div> <!-- PastGrants --> | |||
<div id="TEAM" class="tab-pane fade" role="tabpanel"> | |||
<br/> | <br/> | ||
<span style="font-size:x-large;">''Mechanistic aspects and spatial effects in cell signalling''</span> | <span style="font-size:x-large;">''Mechanistic aspects and spatial effects in cell signalling''</span> | ||
| Line 71: | Line 105: | ||
[[Image:Group_photo_TEAM.jpg|right|thumb | [[Image:Group_photo_TEAM.jpg|right|thumb|link={{SERVER}}/images/b/b6/Group_photo_TEAM.jpg|alt=Photo by Magdalena Wiśniewska-Krasińska|TEAM in December 2009.]] | ||
====TEAM project laureates==== | ====TEAM project laureates==== | ||
| Line 92: | Line 126: | ||
====Financed topics==== | ====Financed topics==== | ||
# ''Theoretical modeling and experimental analysis of regulatory pathways connected with innate immune response''. The candidates are expected to have laboratory experience or/and knowledge in the field of mathematical biology, stochastic processes and differential equations | # ''Theoretical modeling and experimental analysis of regulatory pathways connected with innate immune response''. The candidates are expected to have laboratory experience or/and knowledge in the field of mathematical biology, stochastic processes and differential equations.<br/><br/> | ||
# ''Evolution and co-evolution of genes (and their regulatory regions) related to IRF3 and NF-κB pathway''. Such analysis will be helpful in analysis of crosstalk of IRF3 and NF-κB pathways. The | |||
# ''Evolution and co-evolution of genes (and their regulatory regions) related to IRF3 and NF-κB pathway''. Such analysis will be helpful in analysis of crosstalk of IRF3 and NF-κB pathways. The evolution of genes is related to their biological importance, typically well conserved genes play key roles in the regulatory pathway. Transcription factors IRF3 and NF-κB play central role in innate immune responses. The student will have to learn BLAST (Basic Local Alignment Search Tool) and how to use genome databases and clustering tools.<br/><br/> | |||
# ''Mutations of p53 pathways genes in various types of cancer''. Various types of cancers are associated with specific mutations of p53, Mdm2, Akt, PTEN and other regulating genes. The students will have to search databases to find the mutations (or deletions) and analyze what is their potential impact on dynamics of p53 regulatory pathways we consider. | # ''Mutations of p53 pathways genes in various types of cancer''. Various types of cancers are associated with specific mutations of p53, Mdm2, Akt, PTEN and other regulating genes. The students will have to search databases to find the mutations (or deletions) and analyze what is their potential impact on dynamics of p53 regulatory pathways we consider. | ||
| Line 126: | Line 162: | ||
* Michael R.H. White (Liverpool University, UK) | * Michael R.H. White (Liverpool University, UK) | ||
<br /> | <br/> | ||
[[Image:Logo_innovative_economy.jpg|center|frameless|link=]] | [[Image:Logo_innovative_economy.jpg|center|frameless|link=]] | ||
</div> | |||
</div> | </div> <!-- TEAM --> | ||
<div id="GRIEG" class="tab-pane fade" role="tabpanel"> | |||
<ul class="nav nav-pills" id="grieg-tabs" role="tablist"> | |||
<li class="nav-item"><btn data-toggle="tab" class="nav-link active" role="tab" aria-controls="grieg-subtab-en" aria-selected="true">#grieg-subtab-en|English</btn></li> | |||
<li class="nav-item"><btn data-toggle="tab" class="nav-link" role="tab" aria-controls="tab-pl" aria-selected="false">#grieg-subtab-pl|Polski</btn></li> | |||
</ul> | |||
<div class="tab-content"> | |||
<div id="grieg-subtab-en" class="tab-pane fade show active" role="tabpanel"> | |||
<br/> | |||
[[File:Logo_Norway_Grants.png|88px|left|link=|alt=]] | |||
<span style="font-size:x-large;">''Interaction of natural killer cells with infected cancer cell population: microfluidics-based experiments and single-cell-level mathematical modeling''</span> | |||
===Concepts and objectives=== | |||
The aim of the project is to analyze interactions of natural killer (NK) cells with an infected epithelial cell population at the single-cell resolution. We investigate two respiratory epithelial cell lines: cancerous and non-cancerous, and two respiratory viruses: respiratory syncytial virus (RSV) and influenza A virus (IAV). Our main objectives are as follows: | |||
* Elucidation of the impact of the NK cell-induced cell death on the dynamics of propagation and eradication of RSV and IAV infections. | |||
* Determination of the potential of NK cells to induce immunogenic cell death in infected cells, that can potentially activate adaptive immune response. | |||
* Development of a droplet-based microfluidic system to perform experiments on co-cultures of epithelial and NK cells (living in droplets), that will enable data acquisition at the single-cell resolution. | |||
* Development of a mathematical model capturing interactions of NK cells with an infected cell population at the single-cell resolution. | |||
===Team=== | |||
[[File:Tomasz_Lipniacki.jpg|150px|right|alt=|Prof. Tomasz Lipniacki|link=|frameless]] | |||
IPPT PAN: | |||
* '''Prof. Tomasz Lipniacki, PhD''' (head) | |||
* Sławomir Błoński, PhD | |||
* Maciej Czerkies, PhD | |||
* Frederic Grabowski, BSc | |||
* Joanna Jaruszewicz-Błońska, PhD | |||
* Marek Kochańczyk, PhD | |||
* Piotr Korczyk, PhD | |||
* Zbigniew Korwek, PhD | |||
* Paulina Koza, PhD | |||
* Barbara Kupikowska-Stobba, PhD | |||
* Felipe Nieves Marques Porto, MSc | |||
* Wiktor Prus, PhD | |||
* Damian Zaremba, MSc | |||
SINTEF AS (core team): | |||
* Elizaveta Vereshchagina, PhD (head) | |||
* Linda Sønstevold, PhD | |||
* Michal Marek Mielnik, PhD | |||
* Geir Uri Jensen, PhD | |||
* Shruti Jain, MSc | |||
* Enrique Escobedo-Cousin, PhD | |||
===Research=== | |||
====Scientific publications==== | |||
* <span class="pmbm">Grabowski F</span>, <span class="pmbm">Kochańczyk M</span>, <span class="pmbm">Korwek Z</span>, <span class="pmbm">Czerkies M</span>, <span class="pmbm">Prus W</span>, <span class="pmbm">Lipniacki T</span>. '''Antagonism between viral infection and innate immunity at the single-cell level''', ''PLOS Pathog'' <u>19</u>(9):e1011597 (2023) [https://pubmed.ncbi.nlm.nih.gov/37669278 PubMed] [https://doi.org/10.1371/journal.ppat.1011597 CrossRef] | [https://doi.org/10.1101/2022.11.18.517110 bioRxiv] [https://github.com/grfrederic/visavis Code] [https://doi.org/10.5281/zenodo.7428925 Data]<br/><br/> | |||
* <span class="pmbm">Kurniawan T</span>, <span class="nopmbm">Sahebdivani M</span>, <span class="pmbm">Zaremba D</span>, <span class="pmbm">Blonski S</span>, <span class="nopmbm">Garstecki P</span>, <span class="nopmbm">van Steijn V</span>, <span class="pmbm">Korczyk PM</span>. '''Formation of droplets in microfluidic cross-junctions at small capillary numbers: Breakdown of the classical squeezing regime''', ''Chem Eng J'' <u>474</u>:145601 (2023) [https://doi.org/10.1016/j.cej.2023.145601 CrossRef]<br/><br/> | |||
* <span class="pmbm">Grabowski F</span>, <span class="pmbm">Nałęcz-Jawecki P</span>, <span class="pmbm">Lipniacki T</span>. '''Predictive power of non-identifiable models''', ''Sci Rep'' <u>13</u>:11143 (2023) [https://doi.org/10.1038/s41598-023-37939-8 CrossRef] [https://doi.org/10.1101/2023.04.07.536025 bioRxiv] [https://github.com/grfrederic/identifiability Code]<br/><br/> | |||
* <span class="nopmbm">Sønstevold L</span>, <span class="pmbm">Czerkies M</span>, <span class="nopmbm">Escobedo-Cousin E</span>, <span class="pmbm">Blonski S</span>, <span class="nopmbm">Vereshchagina E</span>. '''Application of polymethylpentene, an oxygen permeable thermoplastic, for long-term on-a-chip cell culture and organ-on-a-chip devices''', ''Micromachines'' <u>14</u>(3):532 (2023) [https://dx.doi.org/10.3390/mi14030532 CrossRef]<br/><br/> | |||
* <span class="author">Czerkies M</span>, <span class="author">Kochańczyk M</span>, <span class="author">Korwek Z</span>, <span class="author">Prus W</span>, <span class="author">Lipniacki T</span>. '''Respiratory syncytial virus protects bystander cells against influenza A virus infection by triggering secretion of type I and type III interferons''', ''J Virol'' (2022) <u>96</u>:22 e01341-22 [https://doi.org/10.1128/jvi.01341-22 CrossRef] [https://doi.org/10.1101/2021.10.11.463877 bioRxiv]<br/>[[File:article_RSV_IAV_crossprotection.png|450px|link=https://doi.org/10.1128/jvi.01341-22]]<br/><br/> | |||
* <span class="pmbm">Grabowski F</span>, <span class="nopmbm">Preibisch G</span>, <span class="nopmbm">Giziński S</span>, <span class="pmbm">Kochańczyk M</span>, <span class="pmbm">Lipniacki T</span>. '''SARS-CoV-2 Variant of Concern 202012/01 has about twofold replicative advantage and acquires concerning mutations''', ''Viruses'' <u>13</u>(3):392 (2021) [https://doi.org/10.3390/v13030392 CrossRef] [https://doi.org/10.1101/2020.12.28.20248906 medRxiv] | [https://www.mdpi.com/1999-4915/13/3/392/pdf PDF]<br/>[[File:article_alpha_1.png|x175px|link=https://doi.org/10.3390/v13030392]] [[File:article_alpha_2.png|x175px|link=https://doi.org/10.3390/v13030392]]<br/><br/> | |||
* <span class="pmbm">Grabowski F</span>, <span class="pmbm">Kochańczyk M</span>, <span class="pmbm">Lipniacki T</span>. '''The spread of SARS-CoV-2 variant Omicron with the doubling time of 2.0–3.3 days can be explained by immune evasion''', ''Viruses'' <u>14</u>(2):294 (2022) [https://doi.org/10.3390/v14020294 CrossRef] [https://dx.doi.org/10.1101/2021.12.08.21267494 medRxiv] | [https://www.mdpi.com/1999-4915/14/2/294/pdf PDF] [https://susy.mdpi.com/user/review/displayFile/23340271/tDGR6nk0?file=author-coverletter&report=16624478 Correspondence]<br/>[[File:article_omicron_1.png|x175px|link=https://doi.org/10.3390/v14020294]]<br/><br/> | |||
* <span class="nopmbm">Mines RC</span>, <span class="pmbm">Lipniacki T</span>, <span class="nopmbm">Shen X</span>. '''Slow nucleosome dynamics set the transcriptional speed limit and induce RNA polymerase II traffic jams and bursts''', ''PLOS Comput Biol'' <u>18</u>(2):e1009811 (2022) [https://doi.org/10.1371/journal.pcbi.1009811 CrossRef]<br/>[[File:article_nucleodyn_1.png|175px|link=https://doi.org/10.1371/journal.pcbi.1009811]]<br/><br/> | |||
* <span class="pmbm">Korwek Z</span>, <span class="pmbm">Czerkies M</span>, <span class="pmbm">Jaruszewicz-Błońska J</span>, <span class="pmbm">Prus W</span>, <span class="pmbm">Kosiuk I</span>, <span class="pmbm">Kochańczyk M</span>, <span class="pmbm">Lipniacki T</span>. '''Non-self RNA rewires IFNβ signaling: A mathematical model of the innate immune response''' (accepted at ''Sci Signal'') [https://doi.org/10.1101/2022.01.30.478391 bioRxiv]<br/>[[File:article_non-self_1.png|300px|link=https://doi.org/10.1101/2022.01.30.478391]]<br/>[[File:article_non-self_2.png|300px|link=https://doi.org/10.1101/2022.01.30.478391]]<br/><br/> | |||
====Other==== | |||
* Lecture: ''SARS-CoV-2 super-spreaders and super-spreading Variant of Concern 202012/01'', University of Warsaw, Faculty of Physics (Tomasz Lipniacki) | |||
* Lecture: ''SARS‐CoV‐2 Variant of Concern 202012/01 has about twofold replicative advantage and acquires concerning mutations'', University of Warsaw, Faculty of Mathematics, Informatics and Mechanics, BOB seminar (Frederic Grabowski) | |||
* Keynote lecture: ''Mathematical modeling of innate immune response to viral infection'', Joint KMM-VIN / ViCEM / ESB cross-disciplinary workshop, Vienna, Austria 2022 (Tomasz Lipniacki) [https://www.conftool.com/kmm-vicem-esb-2022/sessions.php Conference website] | |||
* Talk: ''Dissecting innate immunity responses to viral infection at the single-cell level'', Inverse Problems Meeting (virtual conference) [https://sites.google.com/view/inverseproblems/speakers#h.j2k1s8my7uft Video] | |||
* Talk: ''Non-self RNA rewires IFNβ signaling: A model of the innate immune response'', q-Bio 2022, Colorado State University (Tomasz Lipniacki) [https://q-bio.org/wp/wp-content/uploads/2022/05/Lipniacki_Tomasz.pdf Abstract] [https://pmbm.ippt.pan.pl/presentations/2022--q-Bio--Lipniacki.pdf Slides] | |||
===Technology=== | |||
With our collaborators from SINTEF AS we investigate the performance of the thermoplastic material PMP in microfluidic devices for organ-on-a-chip and long-term cell cultivation on chip applications. The assessment includes investigating cell adherence and proliferation, material suitability for confocal microscopy, compatibility with common chemicals, device fabrication, and most importantly assessment of gas permeability for oxygen supply during 4 days of cell culturing. | |||
[[File:PMP_device_1.png|x200px]] | |||
[[File:PMP_device_2.png|x200px]] | |||
<br/> | |||
Device used in experiments (article under preparation). | |||
===Dissemination=== | |||
====Press==== | |||
* Gazeta Wyborcza, interview, March 2020 ([https://wyborcza.pl/7,75400,25824141,matematyczny-model-rozwoju-epidemii-prof-lipniacki-ograniczenia.html Polish] | [https://wyborcza.pl/7,173236,25831548,poland-follows-down-the-italian-path-prof-lipniacki-author.html English]) | |||
* Gazeta Wyborcza, interview, June 2020 ([https://wyborcza.pl/7,75400,25920600,prof-lipniacki-musimy-wiedziec-kto-i-jak-sie-zakaza-aby.html Polish]) | |||
* Newsweek Polska, interview, October 2020 ([https://www.newsweek.pl/polska/koronawirus-w-polsce-geometryczny-niemal-wzrost-zachorowan-na-covid-19-co-dalej/dby9xdd Polish]) | |||
* Gazeta Wyborcza, article, October 2020 ([https://wyborcza.pl/7,75400,26410669,prof-lipniacki-lockdown-tak-ostry-i-natychmiast.html Polish]) | |||
* Gazeta Wyborcza, article, November 2020 ([https://wyborcza.pl/7,75400,26503391,prof-lipniacki-polska-leci-w-covidowej-mgle.html Polish]) | |||
* Gazeta Wyborcza, interview, July 2021 ([https://wyborcza.pl/7,75400,26665472,prof-lipniacki-sars-cov-2-po-raz-kolejny-znaczaco-zmutowal.html Polish]) | |||
====Broadcast==== | |||
* Polsat News, debate on pandemic development and role of vaccinations, November 2020 ([https://www.youtube.com/watch?v=KQIvgg9Oopo&t=391 Polish]) | |||
* Polsat News, debate on New Year restrictions, December 2020 ([https://www.youtube.com/watch?v=EvZq4QO9CVw&t=75s Polish]) | |||
* TVN24, debate on lockdown strategy, April 2021 ([https://tvn24.pl/polska/koronawirus-w-polsce-profesor-tomasz-lipniacki-i-doktor-habilitowana-barbara-pabjan-komentuja-5065942 Polish]) | |||
* Polsat News, debate on 4th wave of COVID-19, August 2021 ([https://www.youtube.com/watch?v=rFn_J6nDjo0&t=1554s Polish]) | |||
* Radio TOK FM, three separate conversations ([https://audycje.tokfm.pl/gosc/13792,prof-Tomasz-Lipniacki Polish]) | |||
====Public lectures==== | |||
* lecture in German Historical Institute Warsaw ([https://www.dhi.waw.pl/pl/wydarzenia/wyklady/detale/prof-tomasz-lipniacki-virale-invasionen-in-geweben-und-gesellschaften]) | |||
===Contact=== | |||
Lab head – [http://www.ippt.pan.pl/en/staff/tlipnia Prof. Tomasz Lipniacki]. | |||
===Funding=== | |||
[[Image:Logo_Norway_Grants.png|40px|left|link=|alt=]] | |||
This research is funded by the '''Norwegian Financial Mechanism GRIEG-1'''<br/> | |||
(grant 2019/34/H/NZ6/00699, operated by the National Science Centre, Poland). | |||
</div> <!-- grieg-subtab-en --> | |||
<div id="grieg-subtab-pl" class="tab-pane fade" role="tabpanel"> | |||
<br/> | |||
[[File:Logo_Norway_Grants.png|88px|left|link=|alt=]] | |||
<span style="font-size:x-large;">''Oddziaływanie komórek NK z populacją zainfekowanych komórek rakowych: eksperymenty w urządzeniach mikroprzepływowych i modelowanie matematyczne na poziomie pojedynczych komórek''</span> | |||
===Cele projektu=== | |||
Celem projektu jest analiza oddziaływań komórek “Naturalnych Zabójców” (ang. Natural Killer, NK) z zakażonymi rakowymi komórkami nabłonka płuc na poziomie pojedynczych komórek. Badamy dwie linie komórek nabłonka oddechowego, z których jedna ma pochodzenie nowotworowe oraz dwa wirusy: syncytialny wirus nabłonka oddechowego (RSV) oraz wirus grypy typu A (IAV). Nasze główne zadania badawcze są następujące: | |||
* wyjaśnienie wpływu cytotoksyczności komórek NK wobec zakażonych komórek na dynamikę rozprzestrzeniania się oraz eliminacji infekcji wirusami RSV i IAV; | |||
* ustalenie potencjału komórek NK do indukowania w zakażonych komórkach tzw. immunogennej śmierci komórkowej zdolnej do aktywacji mechanizmów nabytej obrony odpornościowej; | |||
* rozwój systemu mikrofluidyki kropelkowej przeznaczonej prowadzenia doświadczeń na kokulturach komórek nabłonkowych i komórek NK, który pozwoli na zbieranie danych na poziomie poszczególnych komórek; | |||
* rozwój modelu matematycznego opisującego interakcje komórek NK z zainfekowaną populacją komórek nabłonkowych, na poziomie pojedynczych komórek. | |||
===Zespół=== | |||
[[Image:Tomasz_Lipniacki.jpg|150px|right|alt=|link=|Prof. Tomasz Lipniacki|frameless]] | |||
IPPT PAN: | |||
* '''prof. Tomasz Lipniacki (kierownik projektu)''' | |||
* dr inż. Sławomir Błoński | |||
* dr Maciej Czerkies | |||
* lic. Frederic Grabowski | |||
* dr Joanna Jaruszewicz-Błońska | |||
* dr Marek Kochańczyk | |||
* dr Piotr Korczyk | |||
* dr Zbigniew Korwek | |||
* dr Paulina Koza | |||
* dr inż. Barbara Kupikowska-Stobba | |||
* mgr Felipe Nieves Marques Porto | |||
* dr inż. Wiktor Prus | |||
* mgr inż. Damian Zaremba | |||
SINTEF AS (core team): | |||
* dr Elizaveta Vereshchagina (koordynator strony norweskiej) | |||
* dr Linda Sønstevold | |||
* dr Michal Marek Mielnik | |||
* dr Geir Uri Jensen | |||
* mgr Shruti Jain | |||
* dr Enrique Escobedo-Cousin | |||
===Wyniki=== | |||
====Publikacje==== | |||
* <span class="pmbm">Grabowski F</span>, <span class="pmbm">Kochańczyk M</span>, <span class="pmbm">Korwek Z</span>, <span class="pmbm">Czerkies M</span>, <span class="pmbm">Prus W</span>, <span class="pmbm">Lipniacki T</span>. '''Antagonism between viral infection and innate immunity at the single-cell level''', ''PLOS Pathog'' <u>19</u>(9):e1011597 (2023) [https://pubmed.ncbi.nlm.nih.gov/37669278 PubMed] [https://doi.org/10.1371/journal.ppat.1011597 CrossRef] | [https://doi.org/10.1101/2022.11.18.517110 bioRxiv] [https://github.com/grfrederic/visavis Code] [https://doi.org/10.5281/zenodo.7428925 Data]<br/><br/> | |||
* <span class="pmbm">Kurniawan T</span>, <span class="nopmbm">Sahebdivani M</span>, <span class="pmbm">Zaremba D</span>, <span class="pmbm">Blonski S</span>, <span class="nopmbm">Garstecki P</span>, <span class="nopmbm">van Steijn V</span>, <span class="pmbm">Korczyk PM</span>. '''Formation of droplets in microfluidic cross-junctions at small capillary numbers: Breakdown of the classical squeezing regime''', ''Chem Eng J'' <u>474</u>:145601 (2023) [https://doi.org/10.1016/j.cej.2023.145601 CrossRef]<br/><br/> | |||
* <span class="pmbm">Grabowski F</span>, <span class="pmbm">Nałęcz-Jawecki P</span>, <span class="pmbm">Lipniacki T</span>. '''Predictive power of non-identifiable models''', ''Sci Rep'' <u>13</u>:11143 (2023) [https://doi.org/10.1038/s41598-023-37939-8 CrossRef] [https://doi.org/10.1101/2023.04.07.536025 bioRxiv] [https://github.com/grfrederic/identifiability Code]<br/><br/> | |||
* <span class="nopmbm">Sønstevold L</span>, <span class="pmbm">Czerkies M</span>, <span class="nopmbm">Escobedo-Cousin E</span>, <span class="pmbm">Blonski S</span>, <span class="nopmbm">Vereshchagina E</span>. '''Application of polymethylpentene, an oxygen permeable thermoplastic, for long-term on-a-chip cell culture and organ-on-a-chip devices''', ''Micromachines'' <u>14</u>(3):532 (2023) [https://dx.doi.org/10.3390/mi14030532 CrossRef]<br/><br/> | |||
* <span class="author">Czerkies M</span>, <span class="author">Kochańczyk M</span>, <span class="author">Korwek Z</span>, <span class="author">Prus W</span>, <span class="author">Lipniacki T</span>. '''Respiratory syncytial virus protects bystander cells against influenza A virus infection by triggering secretion of type I and type III interferons''', ''J Virol'' (2022) <u>96</u>:22 e01341-22 [https://doi.org/10.1128/jvi.01341-22 CrossRef] [https://doi.org/10.1101/2021.10.11.463877 bioRxiv]<br/>[[File:article_RSV_IAV_crossprotection.png|450px|link=https://doi.org/10.1128/jvi.01341-22]]<br/><br/> | |||
* <span class="pmbm">Grabowski F</span>, <span class="nopmbm">Preibisch G</span>, <span class="nopmbm">Giziński S</span>, <span class="pmbm">Kochańczyk M</span>, <span class="pmbm">Lipniacki T</span>. '''SARS-CoV-2 Variant of Concern 202012/01 has about twofold replicative advantage and acquires concerning mutations''', ''Viruses'' <u>13</u>(3):392 (2021) [https://doi.org/10.3390/v13030392 CrossRef] [https://doi.org/10.1101/2020.12.28.20248906 medRxiv] | [https://www.mdpi.com/1999-4915/13/3/392/pdf PDF]<br/>[[File:article_alpha_1.png|x175px|link=https://doi.org/10.3390/v13030392]] [[File:article_alpha_2.png|x175px|link=https://doi.org/10.3390/v13030392]]<br/><br/> | |||
* <span class="pmbm">Grabowski F</span>, <span class="pmbm">Kochańczyk M</span>, <span class="pmbm">Lipniacki T</span>. '''The spread of SARS-CoV-2 variant Omicron with the doubling time of 2.0–3.3 days can be explained by immune evasion''', ''Viruses'' <u>14</u>(2):294 (2022) [https://doi.org/10.3390/v14020294 CrossRef] [https://dx.doi.org/10.1101/2021.12.08.21267494 medRxiv] | [https://www.mdpi.com/1999-4915/14/2/294/pdf PDF] [https://susy.mdpi.com/user/review/displayFile/23340271/tDGR6nk0?file=author-coverletter&report=16624478 Korespondencja]<br/>[[File:article_omicron_1.png|x175px|link=https://doi.org/10.3390/v14020294]]<br/><br/> | |||
* <span class="nopmbm">Mines RC</span>, <span class="pmbm">Lipniacki T</span>, <span class="nopmbm">Shen X</span>. '''Slow nucleosome dynamics set the transcriptional speed limit and induce RNA polymerase II traffic jams and bursts''', ''PLOS Comput Biol'' <u>18</u>(2):e1009811 (2022) [https://doi.org/10.1371/journal.pcbi.1009811 CrossRef]<br/>[[File:article_nucleodyn_1.png|175px|link=https://doi.org/10.1371/journal.pcbi.1009811]]<br/><br/> | |||
* <span class="pmbm">Korwek Z</span>, <span class="pmbm">Czerkies M</span>, <span class="pmbm">Jaruszewicz-Błońska J</span>, <span class="pmbm">Prus W</span>, <span class="pmbm">Kosiuk I</span>, <span class="pmbm">Kochańczyk M</span>, <span class="pmbm">Lipniacki T</span>. '''Non-self RNA rewires IFNβ signaling: A mathematical model of the innate immune response''' (przyjęte do ''Sci Signal'') [https://doi.org/10.1101/2022.01.30.478391 bioRxiv]<br/>[[File:article_non-self_1.png|300px|link=https://doi.org/10.1101/2022.01.30.478391]]<br/>[[File:article_non-self_2.png|300px|link=https://doi.org/10.1101/2022.01.30.478391]]<br/><br/> | |||
====Inne==== | |||
* Wykład: ''SARS-CoV-2 super-spreaders and super-spreading Variant of Concern 202012/01'', Uniwersytet Warszawski, Wydział Fizyki (Tomasz Lipniacki) | |||
* Wykład: ''SARS‐CoV‐2 Variant of Concern 202012/01 has about twofold replicative advantage and acquires concerning mutations'', Uniwersytet Warszawski, Wydział Matematyki, Informatyki i Mechaniki, seminarium BOB (Frederic Grabowski) | |||
* Wykład zaproszony: ''Mathematical modeling of innate immune response to viral infection'', Joint KMM-VIN / ViCEM / ESB cross-disciplinary workshop, Vienna, Austria 2022 (Tomasz Lipniacki) [https://www.conftool.com/kmm-vicem-esb-2022/sessions.php strona konferencji] | |||
* Wystąpienie: ''Dissecting innate immunity responses to viral infection at the single-cell level'', Inverse Problems Meeting (virtual conference) [https://sites.google.com/view/inverseproblems/speakers#h.j2k1s8my7uft dostępne online] | |||
* Wystąpienie: ''Non-self RNA rewires IFNβ signaling: A model of the innate immune response'', q-Bio 2022, Colorado State University (Tomasz Lipniacki) [https://q-bio.org/wp/wp-content/uploads/2022/05/Lipniacki_Tomasz.pdf podsumowanie] [https://pmbm.ippt.pan.pl/presentations/2022--q-Bio--Lipniacki.pdf Slajdy] | |||
===Technologia=== | |||
Wraz z naszymi współpracownikami z SINTEF AS badamy działanie termoplastycznego materiału PMP w urządzeniach mikroprzepływowych do organ-on-a-chip i długoterminowej hodowli komórek na chipach. Ocena obejmuje badanie przylegania i proliferacji komórek, przydatność materiału do mikroskopii konfokalnej, zgodność z powszechnie stosowanymi substancjami chemicznymi, wytwarzanie urządzeń i, co najważniejsze, ocenę przepuszczalności gazu dla dostarczania tlenu podczas 4 dni hodowli komórek. | |||
[[File:PMP_device_1.png|x200px]] | |||
[[File:PMP_device_2.png|x200px]] | |||
<br/> | |||
Urządzenie używane w eksperymentach (publikacja w przygotowaniu). | |||
===Popularyzacja=== | |||
====Prasa==== | |||
* Gazeta Wyborcza, wywiad, marzec 2020 ([https://wyborcza.pl/7,75400,25824141,matematyczny-model-rozwoju-epidemii-prof-lipniacki-ograniczenia.html po polsku], [https://wyborcza.pl/7,173236,25831548,poland-follows-down-the-italian-path-prof-lipniacki-author.html po angielsku]) | |||
* Gazeta Wyborcza, wywiad, czerwiec 2020 ([https://wyborcza.pl/7,75400,25920600,prof-lipniacki-musimy-wiedziec-kto-i-jak-sie-zakaza-aby.html po polsku]) | |||
* Newsweek Polska, wywiad, październik 2020 ([https://www.newsweek.pl/polska/koronawirus-w-polsce-geometryczny-niemal-wzrost-zachorowan-na-covid-19-co-dalej/dby9xdd po polsku]) | |||
* Gazeta Wyborcza, artykuł, październik 2020 ([https://wyborcza.pl/7,75400,26410669,prof-lipniacki-lockdown-tak-ostry-i-natychmiast.html po polsku]) | |||
* Gazeta Wyborcza, artykuł, listopad 2020 ([https://wyborcza.pl/7,75400,26503391,prof-lipniacki-polska-leci-w-covidowej-mgle.html po polsku]) | |||
* Gazeta Wyborcza, wywiad, lipiec 2021 ([https://wyborcza.pl/7,75400,26665472,prof-lipniacki-sars-cov-2-po-raz-kolejny-znaczaco-zmutowal.html po polsku]) | |||
====Radio i Telewizja==== | |||
* Polsat News, debata o rozwoju pandemii i roli szczepień, listopad 2020 ([https://www.youtube.com/watch?v=KQIvgg9Oopo&t=391 po polsku]) | |||
* Polsat News, debata o restrykcjach sylwestrowych, grudzień 2020 ([https://www.youtube.com/watch?v=EvZq4QO9CVw&t=75s po polsku]) | |||
* TVN24, debata o strategii lockdownów, kwiecień 2021 ([https://tvn24.pl/polska/koronawirus-w-polsce-profesor-tomasz-lipniacki-i-doktor-habilitowana-barbara-pabjan-komentuja-5065942 po polsku]) | |||
* Polsat News, debata o czwartej fali COVID-19, sierpień 2021 ([https://www.youtube.com/watch?v=rFn_J6nDjo0&t=1554s po polsku]) | |||
* Radio TOK FM, trzy rozmowy ([https://audycje.tokfm.pl/gosc/13792,prof-Tomasz-Lipniacki po polsku]) | |||
===Inne=== | |||
* wykład w Niemieckim Instytucie Historycznym w Warszawie ([https://www.dhi.waw.pl/pl/wydarzenia/wyklady/detale/prof-tomasz-lipniacki-virale-invasionen-in-geweben-und-gesellschaften]) | |||
===Kontakt=== | |||
Kierownik Pracowni – [http://www.ippt.pan.pl/en/staff/tlipnia Prof. Tomasz Lipniacki]. | |||
===Finansowanie=== | |||
[[Image:Logo_Norway_Grants.png|40px|left|link=|alt=]] | |||
Badania są finansowane z '''Norweskiego Mechanizmu Finansowego GRIEG-1'''<br/> | |||
(grant 2019/34/H/NZ6/00699, którego operatorem jest Narodowe Centrum Nauki). | |||
</div> <!-- grieg-subtab-pl --> | |||
</div> <!-- tab-content (inner - language for GRIEG)--> | |||
</div> <!-- tab-content (outer) --> | |||
__NOTOC__ | __NOTOC__ | ||
Latest revision as of 20:50, 22 October 2023
- Combinatorial targeting of non-genetic sources of cell-to-cell heterogeneity to overcome fractional killing of cancer cells.
National Science Centre (NCN): Opus, 2022–2025.
PI: Jarosław Walczak
- Cell-to-cell heterogeneity of cross-wired cytokine signaling: filling the gaps between molecular origins and translational implications.
National Science Centre (NCN): Opus, 2021–2025.
PI: Michał Komorowski
- Interaction of natural killer cells with an infected cancer cell population: microfluidics-based experiments and single-cell-level mathematical modeling.
National Science Centre (NCN): Grieg (Norwegian Financial Mechanism), 2020–2023.
PI: Tomasz Lipniacki
- Influence of oncogenic mutations on information transmission in the MAPK signaling pathway.
National Science Centre (NCN): Opus, 2020–2024.
PI: Tomasz Lipniacki
- Sprayable multifunctional nanofibrous hydrogels for polytherapeutic wound healing treatments.
National Centre for Research and Development (NCBiR): 2nd Polish–Chinese/Chinese–Polish Joint Research Call, 2021–2024.
PI: Filippo Pierini
- Electrospun conducting hydrogel nanomaterials for neural tissue engineering.
Foundation for Polish Science (FNP): First Team, 2019–2022.
PI: Filippo Pierini
- Coordination of innate immune response in infected cell population: experiment and mathematical modeling.
National Science Centre (NCN): Opus 15, 2019–2022.
PI: Tomasz Lipniacki
- Deciphering biochemical signalling to inform more efficient therapeutic strategies.
Foundation for Polish Science (FNP): First Team, 2018–2021
PI: Michał Komorowski
- Development of novel methods for measuring viscosity and interfacial tension of liquids using microfluidic devices.
National Science Centre (NCN): Preludium 15, 2019–2022.
PI: Damian Zaremba
- Exlaining intercellular heterogeneity of the response to IFNγ.
National Science Centre (NCN): Preludium 17, 2020–2022.
PI: Piotr Topolewski
- Injectible cell scaffolds in tissue engineering.
National Centre for Research and Development (NCBiR): Lider 9, 2019–2022.
PI: Paweł Nakielski
- Self-organization and nano-mechanical characterization of cellulose microfibers.
National Science Centre (NCN): Sonata 13, 2018–2021.
PI: Adolfo Poma Bernaola
- Digital operations on droplets embedded into smart microfluidic architectures for applications in medical diagnostics and biological research.
Foundation for Polish Science (FNP): First Team, 2018–2020.
PI: Piotr Korczyk
- Design, construction and optimization of a combined Atomic Force Microscope and Optical Tweezers instrument for single molecules and nanomaterials characterization.
National Centre for Research and Development (NCBiR): Lider 7, 2017–2020.
PI: Filippo Pierini
- The effect of shear flow on fibrin clot structure and fibrin-platelets interactions.
Foundation for Polish Science (FNP): Homing, 2017–2020.
PI: Izabela Piechocka
- Non-standard parabolic problems in description of biological processes.
National Science Centre (NCN): OPUS 11, 2017–2020.
PI: Bogdan Kaźmierczak
- Innate immune response to RSV infection, role of non-structural proteins. Experiments and modeling of signal transduction and among cells communication.
National Science Centre (NCN): Harmonia 6, 2015–2019.
PI: Tomasz Lipniacki
- Stimuli-responsive chiral nematic liquid crystal hydrogel implants by electrospinning technique.
National Science Centre (NCN): Sonata 10, 2016–2019.
PI: Filippo Pierini
- Investigation of blood clotting mechanism in the contact with nanofibers.
National Science Centre (NCN): Sonata 10, 2016–2019.
PI: Paweł Nakielski
- Multi-scale dynamics of signal transduction: dissecting the MAPK pathway.
Wiener Wissenschafts-, Forschungs- und Technologiefonds (WWTF Austria) grant "Mathematics and...", 2015–2018.
Co-PI: Tomasz Lipniacki
- Dynamics of STAT1, STAT3, and STAT5 phosphorylation in breast cancer cell lines: a systematic theoretical and experimental analysis.
National Science Centre (NCN): Opus 9, 2016–2019.
PI: Michał Komorowski
- Detecting signalling aberrations in cancer cells.
National Science Centre (NCN): Preludium 12, 2017–2019.
PI: Karol Nienałtowski
- Analysis of hydrodynamic interactions between single molecules using optical tweezers.
National Science Centre (NCN): Preludium 10, 2016–2019.
PI: Krzysztof Zembrzycki
- Mechanisms controlling cell fate decisions in response to stress. Experimental and theoretical analysis.
National Science Centre (NCN): Opus 7, 2015–2018.
PI: Tomasz Lipniacki.
- The study of hydrodynamic interactions of droplets in complex microfluidic structures. The analysis of algorithms encoded in the architecture of microchannels enabling dosing of the reactants with arbitrary precision.
National Science Centre (NCN): Sonata Bis 4, 2015–2018.
PI: Piotr Korczyk
- European Molecular Biology Organisation (EMBO): Installation Grant, 2013–2016/2018.
PI: Michał Komorowski
- Hydrogel noanofilaments for biomedical application.
National Science Centre (NCN): Preludium 9, 2016–2018.
PI: Sylwia Pawłowska
- Innate immune signalling: optimal microfluidic protocols, prediction and control.
Marie Curie Actions: Career Integration Grant, 2013–2017.
PI: Michał Komorowski.
- Stochastic phenotype switching in growing and dividing bacteria.
National Science Centre (NCN): Preludium 7, 2015–2017.
PI: Joanna Jaruszewicz–Błońska
- Regulation of the RAF/MEK/ERK cascade involving RAF isoforms.
National Science Centre (NCN): Preludium 6, 2014–2017.
PI: Paweł Kocieniewski
- Dynamics of micro and nano objects suspended in fluids.
National Science Centre (NCN): OPUS 2, 2012–2016.
PI: Tomasz Kowalewski
- Formation of spatial heterogeneities on the plasma membrane and their impact on cell signaling.
National Science Centre (NCN): Preludium 5, 2014–2016.
PI: Marek Kochańczyk
- Inter- and intracellular signaling in innate immune response: experimental and mathematical analysis.
National Science Centre (NCN): OPUS 2, 2012–2015.
PI: Tomasz Lipniacki
- Estimation of the kinetic rates in the NF-κΒ signalling system.
Polish Ministry of Science and Higher Education: Iuventus Plus, 2013–2015.
PI: Michał Komorowski
- Mechanistic aspects and spatial effects in cell signaling.
Foundation for Polish Science (FNP): TEAM, 2009–2013.
PI: Tomasz Lipniacki
- Multiparameter and information theoretic models of biochemical signal transduction – modeling and inference.
Foundation for Polish Science (FNP): HOMING Plus, 2011–2013.
PI: Michał Komorowski
- Application of electrospun nanofibrous mats as active wound dressing for prevention of post-traumatic brain damage (Zastosowanie elektroprzędzonych nanowłókien jako opatrunków aktywnych w zapobieganiu pourazowym zmianom w tkance mózgowej).
National Center for Research and Development (NCBiR), 2010–2013.
PI: Tomasz Kowalewski
- Traveling waves in cylindrical domains (Fale biegnące w obszarach cylindrycznych). National Science Centre (NCN), 2010–2012.
PI: Bogdan Kaźmierczak
- Application of information theory-based computational methods for the analysis of signal transduction efficiency in neuronal networks (Zastosowanie metod komputerowych bazujących na teorii informacji do analizy efektywności transmisji sygnałów w sieciach neuronowych).
National Science Centre (NCN), 2011–2012.
PI: Janusz Szczepański
- Efficient methods for generation of secure parameters of public key algorithms (Opracowanie wydajnych metod generowania bezpiecznych parametrów algorytmów klucza publicznego).
National Science Centre (NCN): grant for a Ph.D. student, 2011–2012.
PI: Janusz Szczepański
Mechanistic aspects and spatial effects in cell signalling
within the TEAM Programme sponsored by the Foundation for Polish Science, 2009–2013
Concepts and objectives
The leading aim of the project is to identify and study the turning points in molecular pathways of immune response and cancer, in which the single cell fate, understood as a choice between apoptosis, senescence, proliferation, differentiation or cell cycle arrest, is decided. As a model molecular pathways we will consider these related to innate and adaptive immune response and cancer, in which we have expertise. The project will be realized both by means of numerical simulations and theoretical analysis.
TEAM project laureates
Postdocs:
- Sławomir Błoński
- Piotr Szopa (till 31.01.2012)
Ph.D. Students:
- Michał Dyzma
- Joanna Jaruszewicz
- Paweł Kocieniewski
M.Sc. Students:
- Marta Bogdał
- Dominika Nowicka
- Jakub Pękalski (till 31.08.2011)
- Paweł Żuk (till 30.09.2011)
Financed topics
- Theoretical modeling and experimental analysis of regulatory pathways connected with innate immune response. The candidates are expected to have laboratory experience or/and knowledge in the field of mathematical biology, stochastic processes and differential equations.
- Evolution and co-evolution of genes (and their regulatory regions) related to IRF3 and NF-κB pathway. Such analysis will be helpful in analysis of crosstalk of IRF3 and NF-κB pathways. The evolution of genes is related to their biological importance, typically well conserved genes play key roles in the regulatory pathway. Transcription factors IRF3 and NF-κB play central role in innate immune responses. The student will have to learn BLAST (Basic Local Alignment Search Tool) and how to use genome databases and clustering tools.
- Mutations of p53 pathways genes in various types of cancer. Various types of cancers are associated with specific mutations of p53, Mdm2, Akt, PTEN and other regulating genes. The students will have to search databases to find the mutations (or deletions) and analyze what is their potential impact on dynamics of p53 regulatory pathways we consider.
Team members
- Tomasz Lipniacki – principal investigator (Institute of Fundamental Technological Research, PAS, Warsaw, Poland)
- Bogdan Kaźmierczak (Institute of Fundamental Technological Research, PAS, Warsaw, Poland)
- Beata Hat-Plewińska (Institute of Fundamental Technological Research, PAS, Warsaw, Poland)
- Marek Kochańczyk (Jagiellonian University, Krakow, Poland)
- Aleksandra Nowicka (Institute of Theoretical Physics, PAS, Warsaw, Poland) – till 01.10.2011
External collaborators
- Mark Alber (Notre Dame University, US)
- Allan R. Brasier (University of Texas Medical Branch, Galveston, TX, US)
- James R. Faeder (University of Pittsburgh School of Medicine, US)
- William S. Hlavacek (Los Alamos National Laboratory, US)
- Marek Kimmel (Rice University and MD Anderson Cancer Center, Houston, TX, US)
- Vitaly Volpert (Université Claude Bernard - Lyon 1, France)
- Michael R.H. White (Liverpool University, UK)


Interaction of natural killer cells with infected cancer cell population: microfluidics-based experiments and single-cell-level mathematical modeling
Concepts and objectives
The aim of the project is to analyze interactions of natural killer (NK) cells with an infected epithelial cell population at the single-cell resolution. We investigate two respiratory epithelial cell lines: cancerous and non-cancerous, and two respiratory viruses: respiratory syncytial virus (RSV) and influenza A virus (IAV). Our main objectives are as follows:
- Elucidation of the impact of the NK cell-induced cell death on the dynamics of propagation and eradication of RSV and IAV infections.
- Determination of the potential of NK cells to induce immunogenic cell death in infected cells, that can potentially activate adaptive immune response.
- Development of a droplet-based microfluidic system to perform experiments on co-cultures of epithelial and NK cells (living in droplets), that will enable data acquisition at the single-cell resolution.
- Development of a mathematical model capturing interactions of NK cells with an infected cell population at the single-cell resolution.
Team

IPPT PAN:
- Prof. Tomasz Lipniacki, PhD (head)
- Sławomir Błoński, PhD
- Maciej Czerkies, PhD
- Frederic Grabowski, BSc
- Joanna Jaruszewicz-Błońska, PhD
- Marek Kochańczyk, PhD
- Piotr Korczyk, PhD
- Zbigniew Korwek, PhD
- Paulina Koza, PhD
- Barbara Kupikowska-Stobba, PhD
- Felipe Nieves Marques Porto, MSc
- Wiktor Prus, PhD
- Damian Zaremba, MSc
SINTEF AS (core team):
- Elizaveta Vereshchagina, PhD (head)
- Linda Sønstevold, PhD
- Michal Marek Mielnik, PhD
- Geir Uri Jensen, PhD
- Shruti Jain, MSc
- Enrique Escobedo-Cousin, PhD
Research
Scientific publications
- Grabowski F, Kochańczyk M, Korwek Z, Czerkies M, Prus W, Lipniacki T. Antagonism between viral infection and innate immunity at the single-cell level, PLOS Pathog 19(9):e1011597 (2023) PubMed CrossRef | bioRxiv Code Data
- Kurniawan T, Sahebdivani M, Zaremba D, Blonski S, Garstecki P, van Steijn V, Korczyk PM. Formation of droplets in microfluidic cross-junctions at small capillary numbers: Breakdown of the classical squeezing regime, Chem Eng J 474:145601 (2023) CrossRef
- Grabowski F, Nałęcz-Jawecki P, Lipniacki T. Predictive power of non-identifiable models, Sci Rep 13:11143 (2023) CrossRef bioRxiv Code
- Sønstevold L, Czerkies M, Escobedo-Cousin E, Blonski S, Vereshchagina E. Application of polymethylpentene, an oxygen permeable thermoplastic, for long-term on-a-chip cell culture and organ-on-a-chip devices, Micromachines 14(3):532 (2023) CrossRef
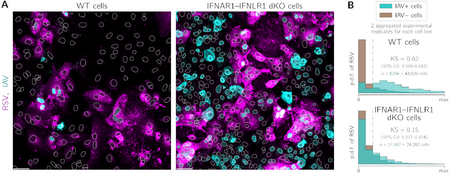
- , , , , . Respiratory syncytial virus protects bystander cells against influenza A virus infection by triggering secretion of type I and type III interferons, J Virol (2022) 96:22 e01341-22 CrossRef bioRxiv
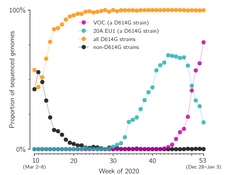
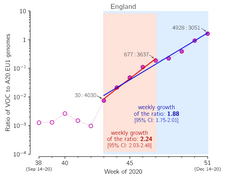
- Grabowski F, Preibisch G, Giziński S, Kochańczyk M, Lipniacki T. SARS-CoV-2 Variant of Concern 202012/01 has about twofold replicative advantage and acquires concerning mutations, Viruses 13(3):392 (2021) CrossRef medRxiv | PDF
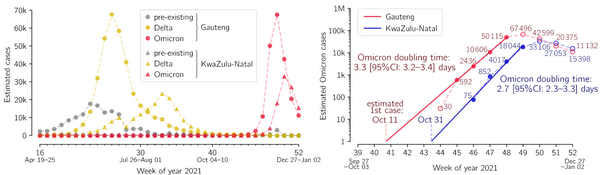
- Grabowski F, Kochańczyk M, Lipniacki T. The spread of SARS-CoV-2 variant Omicron with the doubling time of 2.0–3.3 days can be explained by immune evasion, Viruses 14(2):294 (2022) CrossRef medRxiv | PDF Correspondence
- Mines RC, Lipniacki T, Shen X. Slow nucleosome dynamics set the transcriptional speed limit and induce RNA polymerase II traffic jams and bursts, PLOS Comput Biol 18(2):e1009811 (2022) CrossRef
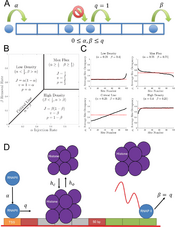
- Korwek Z, Czerkies M, Jaruszewicz-Błońska J, Prus W, Kosiuk I, Kochańczyk M, Lipniacki T. Non-self RNA rewires IFNβ signaling: A mathematical model of the innate immune response (accepted at Sci Signal) bioRxiv
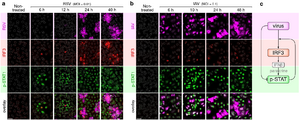
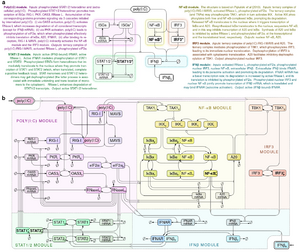
Other
- Lecture: SARS-CoV-2 super-spreaders and super-spreading Variant of Concern 202012/01, University of Warsaw, Faculty of Physics (Tomasz Lipniacki)
- Lecture: SARS‐CoV‐2 Variant of Concern 202012/01 has about twofold replicative advantage and acquires concerning mutations, University of Warsaw, Faculty of Mathematics, Informatics and Mechanics, BOB seminar (Frederic Grabowski)
- Keynote lecture: Mathematical modeling of innate immune response to viral infection, Joint KMM-VIN / ViCEM / ESB cross-disciplinary workshop, Vienna, Austria 2022 (Tomasz Lipniacki) Conference website
- Talk: Dissecting innate immunity responses to viral infection at the single-cell level, Inverse Problems Meeting (virtual conference) Video
- Talk: Non-self RNA rewires IFNβ signaling: A model of the innate immune response, q-Bio 2022, Colorado State University (Tomasz Lipniacki) Abstract Slides
Technology
With our collaborators from SINTEF AS we investigate the performance of the thermoplastic material PMP in microfluidic devices for organ-on-a-chip and long-term cell cultivation on chip applications. The assessment includes investigating cell adherence and proliferation, material suitability for confocal microscopy, compatibility with common chemicals, device fabrication, and most importantly assessment of gas permeability for oxygen supply during 4 days of cell culturing.


Device used in experiments (article under preparation).
Dissemination
Press
- Gazeta Wyborcza, interview, March 2020 (Polish | English)
- Gazeta Wyborcza, interview, June 2020 (Polish)
- Newsweek Polska, interview, October 2020 (Polish)
- Gazeta Wyborcza, article, October 2020 (Polish)
- Gazeta Wyborcza, article, November 2020 (Polish)
- Gazeta Wyborcza, interview, July 2021 (Polish)
Broadcast
- Polsat News, debate on pandemic development and role of vaccinations, November 2020 (Polish)
- Polsat News, debate on New Year restrictions, December 2020 (Polish)
- TVN24, debate on lockdown strategy, April 2021 (Polish)
- Polsat News, debate on 4th wave of COVID-19, August 2021 (Polish)
- Radio TOK FM, three separate conversations (Polish)
Public lectures
- lecture in German Historical Institute Warsaw ([1])
Contact
Lab head – Prof. Tomasz Lipniacki.
Funding
This research is funded by the Norwegian Financial Mechanism GRIEG-1
(grant 2019/34/H/NZ6/00699, operated by the National Science Centre, Poland).

Oddziaływanie komórek NK z populacją zainfekowanych komórek rakowych: eksperymenty w urządzeniach mikroprzepływowych i modelowanie matematyczne na poziomie pojedynczych komórek
Cele projektu
Celem projektu jest analiza oddziaływań komórek “Naturalnych Zabójców” (ang. Natural Killer, NK) z zakażonymi rakowymi komórkami nabłonka płuc na poziomie pojedynczych komórek. Badamy dwie linie komórek nabłonka oddechowego, z których jedna ma pochodzenie nowotworowe oraz dwa wirusy: syncytialny wirus nabłonka oddechowego (RSV) oraz wirus grypy typu A (IAV). Nasze główne zadania badawcze są następujące:
- wyjaśnienie wpływu cytotoksyczności komórek NK wobec zakażonych komórek na dynamikę rozprzestrzeniania się oraz eliminacji infekcji wirusami RSV i IAV;
- ustalenie potencjału komórek NK do indukowania w zakażonych komórkach tzw. immunogennej śmierci komórkowej zdolnej do aktywacji mechanizmów nabytej obrony odpornościowej;
- rozwój systemu mikrofluidyki kropelkowej przeznaczonej prowadzenia doświadczeń na kokulturach komórek nabłonkowych i komórek NK, który pozwoli na zbieranie danych na poziomie poszczególnych komórek;
- rozwój modelu matematycznego opisującego interakcje komórek NK z zainfekowaną populacją komórek nabłonkowych, na poziomie pojedynczych komórek.
Zespół

IPPT PAN:
- prof. Tomasz Lipniacki (kierownik projektu)
- dr inż. Sławomir Błoński
- dr Maciej Czerkies
- lic. Frederic Grabowski
- dr Joanna Jaruszewicz-Błońska
- dr Marek Kochańczyk
- dr Piotr Korczyk
- dr Zbigniew Korwek
- dr Paulina Koza
- dr inż. Barbara Kupikowska-Stobba
- mgr Felipe Nieves Marques Porto
- dr inż. Wiktor Prus
- mgr inż. Damian Zaremba
SINTEF AS (core team):
- dr Elizaveta Vereshchagina (koordynator strony norweskiej)
- dr Linda Sønstevold
- dr Michal Marek Mielnik
- dr Geir Uri Jensen
- mgr Shruti Jain
- dr Enrique Escobedo-Cousin
Wyniki
Publikacje
- Grabowski F, Kochańczyk M, Korwek Z, Czerkies M, Prus W, Lipniacki T. Antagonism between viral infection and innate immunity at the single-cell level, PLOS Pathog 19(9):e1011597 (2023) PubMed CrossRef | bioRxiv Code Data
- Kurniawan T, Sahebdivani M, Zaremba D, Blonski S, Garstecki P, van Steijn V, Korczyk PM. Formation of droplets in microfluidic cross-junctions at small capillary numbers: Breakdown of the classical squeezing regime, Chem Eng J 474:145601 (2023) CrossRef
- Grabowski F, Nałęcz-Jawecki P, Lipniacki T. Predictive power of non-identifiable models, Sci Rep 13:11143 (2023) CrossRef bioRxiv Code
- Sønstevold L, Czerkies M, Escobedo-Cousin E, Blonski S, Vereshchagina E. Application of polymethylpentene, an oxygen permeable thermoplastic, for long-term on-a-chip cell culture and organ-on-a-chip devices, Micromachines 14(3):532 (2023) CrossRef
- , , , , . Respiratory syncytial virus protects bystander cells against influenza A virus infection by triggering secretion of type I and type III interferons, J Virol (2022) 96:22 e01341-22 CrossRef bioRxiv

- Grabowski F, Preibisch G, Giziński S, Kochańczyk M, Lipniacki T. SARS-CoV-2 Variant of Concern 202012/01 has about twofold replicative advantage and acquires concerning mutations, Viruses 13(3):392 (2021) CrossRef medRxiv | PDF


- Grabowski F, Kochańczyk M, Lipniacki T. The spread of SARS-CoV-2 variant Omicron with the doubling time of 2.0–3.3 days can be explained by immune evasion, Viruses 14(2):294 (2022) CrossRef medRxiv | PDF Korespondencja

- Mines RC, Lipniacki T, Shen X. Slow nucleosome dynamics set the transcriptional speed limit and induce RNA polymerase II traffic jams and bursts, PLOS Comput Biol 18(2):e1009811 (2022) CrossRef

- Korwek Z, Czerkies M, Jaruszewicz-Błońska J, Prus W, Kosiuk I, Kochańczyk M, Lipniacki T. Non-self RNA rewires IFNβ signaling: A mathematical model of the innate immune response (przyjęte do Sci Signal) bioRxiv


Inne
- Wykład: SARS-CoV-2 super-spreaders and super-spreading Variant of Concern 202012/01, Uniwersytet Warszawski, Wydział Fizyki (Tomasz Lipniacki)
- Wykład: SARS‐CoV‐2 Variant of Concern 202012/01 has about twofold replicative advantage and acquires concerning mutations, Uniwersytet Warszawski, Wydział Matematyki, Informatyki i Mechaniki, seminarium BOB (Frederic Grabowski)
- Wykład zaproszony: Mathematical modeling of innate immune response to viral infection, Joint KMM-VIN / ViCEM / ESB cross-disciplinary workshop, Vienna, Austria 2022 (Tomasz Lipniacki) strona konferencji
- Wystąpienie: Dissecting innate immunity responses to viral infection at the single-cell level, Inverse Problems Meeting (virtual conference) dostępne online
- Wystąpienie: Non-self RNA rewires IFNβ signaling: A model of the innate immune response, q-Bio 2022, Colorado State University (Tomasz Lipniacki) podsumowanie Slajdy
Technologia
Wraz z naszymi współpracownikami z SINTEF AS badamy działanie termoplastycznego materiału PMP w urządzeniach mikroprzepływowych do organ-on-a-chip i długoterminowej hodowli komórek na chipach. Ocena obejmuje badanie przylegania i proliferacji komórek, przydatność materiału do mikroskopii konfokalnej, zgodność z powszechnie stosowanymi substancjami chemicznymi, wytwarzanie urządzeń i, co najważniejsze, ocenę przepuszczalności gazu dla dostarczania tlenu podczas 4 dni hodowli komórek.


Urządzenie używane w eksperymentach (publikacja w przygotowaniu).
Popularyzacja
Prasa
- Gazeta Wyborcza, wywiad, marzec 2020 (po polsku, po angielsku)
- Gazeta Wyborcza, wywiad, czerwiec 2020 (po polsku)
- Newsweek Polska, wywiad, październik 2020 (po polsku)
- Gazeta Wyborcza, artykuł, październik 2020 (po polsku)
- Gazeta Wyborcza, artykuł, listopad 2020 (po polsku)
- Gazeta Wyborcza, wywiad, lipiec 2021 (po polsku)
Radio i Telewizja
- Polsat News, debata o rozwoju pandemii i roli szczepień, listopad 2020 (po polsku)
- Polsat News, debata o restrykcjach sylwestrowych, grudzień 2020 (po polsku)
- TVN24, debata o strategii lockdownów, kwiecień 2021 (po polsku)
- Polsat News, debata o czwartej fali COVID-19, sierpień 2021 (po polsku)
- Radio TOK FM, trzy rozmowy (po polsku)
Inne
- wykład w Niemieckim Instytucie Historycznym w Warszawie ([2])
Kontakt
Kierownik Pracowni – Prof. Tomasz Lipniacki.
Finansowanie
Badania są finansowane z Norweskiego Mechanizmu Finansowego GRIEG-1
(grant 2019/34/H/NZ6/00699, którego operatorem jest Narodowe Centrum Nauki).

