Grants/Grieg: Difference between revisions
mNo edit summary |
mNo edit summary |
||
| (9 intermediate revisions by the same user not shown) | |||
| Line 5: | Line 5: | ||
<div class="tab-content"> | <div class="tab-content"> | ||
<div id="grieg-subtab-en" class="tab-pane fade | <div id="grieg-subtab-en" class="tab-pane fade show active" role="tabpanel" aria-labelledby="tab1Label"> | ||
<br/> | <br/> | ||
| Line 51: | Line 51: | ||
===Research=== | ===Research=== | ||
====Scientific publications==== | ====Scientific publications==== | ||
* <span class="pmbm">Grabowski F</span>, <span class="pmbm">Kochańczyk M</span>, <span class="pmbm">Korwek Z</span>, <span class="pmbm">Czerkies M</span>, <span class="pmbm">Prus W</span>, <span class="pmbm">Lipniacki T</span>. '''Antagonism between viral infection and innate immunity at the single-cell level''', ''PLOS Pathog'' <u>19</u>(9):e1011597 (2023) [https://pubmed.ncbi.nlm.nih.gov/37669278 PubMed] [https://doi.org/10.1371/journal.ppat.1011597 CrossRef] | [https://doi.org/10.1101/2022.11.18.517110 bioRxiv] [https://github.com/grfrederic/visavis Code] [https://doi.org/10.5281/zenodo.7428925 Data]<br/><br/> | |||
* <span class="pmbm">Kurniawan T</span>, <span class="nopmbm">Sahebdivani M</span>, <span class="pmbm">Zaremba D</span>, <span class="pmbm">Blonski S</span>, <span class="nopmbm">Garstecki P</span>, <span class="nopmbm">van Steijn V</span>, <span class="pmbm">Korczyk PM</span>. '''Formation of droplets in microfluidic cross-junctions at small capillary numbers: Breakdown of the classical squeezing regime''', ''Chem Eng J'' <u>474</u>:145601 (2023) [https://doi.org/10.1016/j.cej.2023.145601 CrossRef]<br/><br/> | |||
* <span class="pmbm">Grabowski F</span>, <span class="pmbm">Nałęcz-Jawecki P</span>, <span class="pmbm">Lipniacki T</span>. '''Predictive power of non-identifiable models''', ''Sci Rep'' <u>13</u>:11143 (2023) [https://doi.org/10.1038/s41598-023-37939-8 CrossRef] [https://doi.org/10.1101/2023.04.07.536025 bioRxiv] [https://github.com/grfrederic/identifiability Code]<br/><br/> | |||
* <span class="nopmbm">Sønstevold L</span>, <span class="pmbm">Czerkies M</span>, <span class="nopmbm">Escobedo-Cousin E</span>, <span class="pmbm">Blonski S</span>, <span class="nopmbm">Vereshchagina E</span>. '''Application of polymethylpentene, an oxygen permeable thermoplastic, for long-term on-a-chip cell culture and organ-on-a-chip devices''', ''Micromachines'' <u>14</u>(3):532 (2023) [https://dx.doi.org/10.3390/mi14030532 CrossRef]<br/><br/> | |||
* <span class="author">Czerkies M</span>, <span class="author">Kochańczyk M</span>, <span class="author">Korwek Z</span>, <span class="author">Prus W</span>, <span class="author">Lipniacki T</span>. '''Respiratory syncytial virus protects bystander cells against influenza A virus infection by triggering secretion of type I and type III interferons''', ''J Virol'' (2022) <u>96</u>:22 e01341-22 [https://doi.org/10.1128/jvi.01341-22 CrossRef] [https://doi.org/10.1101/2021.10.11.463877 bioRxiv]<br/>[[File:article_RSV_IAV_crossprotection.png|450px|link=https://doi.org/10.1128/jvi.01341-22]]<br/><br/> | * <span class="author">Czerkies M</span>, <span class="author">Kochańczyk M</span>, <span class="author">Korwek Z</span>, <span class="author">Prus W</span>, <span class="author">Lipniacki T</span>. '''Respiratory syncytial virus protects bystander cells against influenza A virus infection by triggering secretion of type I and type III interferons''', ''J Virol'' (2022) <u>96</u>:22 e01341-22 [https://doi.org/10.1128/jvi.01341-22 CrossRef] [https://doi.org/10.1101/2021.10.11.463877 bioRxiv]<br/>[[File:article_RSV_IAV_crossprotection.png|450px|link=https://doi.org/10.1128/jvi.01341-22]]<br/><br/> | ||
* <span class="pmbm">Grabowski F</span>, <span class="nopmbm">Preibisch G</span>, <span class="nopmbm">Giziński S</span>, <span class="pmbm">Kochańczyk M</span>, <span class="pmbm">Lipniacki T</span>. '''SARS-CoV-2 Variant of Concern 202012/01 has about twofold replicative advantage and acquires concerning mutations''', ''Viruses'' <u>13</u>(3):392 (2021) [https://doi.org/10.3390/v13030392 CrossRef] [https://doi.org/10.1101/2020.12.28.20248906 medRxiv] | [https://www.mdpi.com/1999-4915/13/3/392/pdf PDF]<br/>[[File:article_alpha_1.png|x175px|link=https://doi.org/10.3390/v13030392]] [[File:article_alpha_2.png|x175px|link=https://doi.org/10.3390/v13030392]]<br/><br/> | * <span class="pmbm">Grabowski F</span>, <span class="nopmbm">Preibisch G</span>, <span class="nopmbm">Giziński S</span>, <span class="pmbm">Kochańczyk M</span>, <span class="pmbm">Lipniacki T</span>. '''SARS-CoV-2 Variant of Concern 202012/01 has about twofold replicative advantage and acquires concerning mutations''', ''Viruses'' <u>13</u>(3):392 (2021) [https://doi.org/10.3390/v13030392 CrossRef] [https://doi.org/10.1101/2020.12.28.20248906 medRxiv] | [https://www.mdpi.com/1999-4915/13/3/392/pdf PDF]<br/>[[File:article_alpha_1.png|x175px|link=https://doi.org/10.3390/v13030392]] [[File:article_alpha_2.png|x175px|link=https://doi.org/10.3390/v13030392]]<br/><br/> | ||
* <span class="pmbm">Grabowski F</span>, <span class="pmbm">Kochańczyk M</span>, <span class="pmbm">Lipniacki T</span>. '''The spread of SARS-CoV-2 variant Omicron with the doubling time of 2.0–3.3 days can be explained by immune evasion''', ''Viruses'' <u>14</u>(2):294 (2022) [https://doi.org/10.3390/v14020294 CrossRef] [https://dx.doi.org/10.1101/2021.12.08.21267494 medRxiv] | [https://www.mdpi.com/1999-4915/14/2/294/pdf PDF] [https://susy.mdpi.com/user/review/displayFile/23340271/tDGR6nk0?file=author-coverletter&report=16624478 Correspondence]<br/>[[File:article_omicron_1.png|x175px|link=https://doi.org/10.3390/v14020294]]<br/><br/> | * <span class="pmbm">Grabowski F</span>, <span class="pmbm">Kochańczyk M</span>, <span class="pmbm">Lipniacki T</span>. '''The spread of SARS-CoV-2 variant Omicron with the doubling time of 2.0–3.3 days can be explained by immune evasion''', ''Viruses'' <u>14</u>(2):294 (2022) [https://doi.org/10.3390/v14020294 CrossRef] [https://dx.doi.org/10.1101/2021.12.08.21267494 medRxiv] | [https://www.mdpi.com/1999-4915/14/2/294/pdf PDF] [https://susy.mdpi.com/user/review/displayFile/23340271/tDGR6nk0?file=author-coverletter&report=16624478 Correspondence]<br/>[[File:article_omicron_1.png|x175px|link=https://doi.org/10.3390/v14020294]]<br/><br/> | ||
* <span class="nopmbm">Mines RC</span>, <span class="pmbm">Lipniacki T</span>, <span class="nopmbm">Shen X</span>. '''Slow nucleosome dynamics set the transcriptional speed limit and induce RNA polymerase II traffic jams and bursts''', ''PLOS Comput Biol'' <u>18</u>(2):e1009811 (2022) [https://doi.org/10.1371/journal.pcbi.1009811 CrossRef]<br/>[[File:article_nucleodyn_1.png|175px|link=https://doi.org/10.1371/journal.pcbi.1009811]]<br/><br/> | * <span class="nopmbm">Mines RC</span>, <span class="pmbm">Lipniacki T</span>, <span class="nopmbm">Shen X</span>. '''Slow nucleosome dynamics set the transcriptional speed limit and induce RNA polymerase II traffic jams and bursts''', ''PLOS Comput Biol'' <u>18</u>(2):e1009811 (2022) [https://doi.org/10.1371/journal.pcbi.1009811 CrossRef]<br/>[[File:article_nucleodyn_1.png|175px|link=https://doi.org/10.1371/journal.pcbi.1009811]]<br/><br/> | ||
* <span class="pmbm">Korwek Z</span>, <span class="pmbm">Czerkies M</span>, <span class="pmbm">Jaruszewicz-Błońska J</span>, <span class="pmbm">Prus W</span>, <span class="pmbm">Kosiuk I</span>, <span class="pmbm">Kochańczyk M</span>, <span class="pmbm">Lipniacki T</span>. '''Non-self RNA rewires IFNβ signaling: A mathematical model of the innate immune response''' ( | * <span class="pmbm">Korwek Z</span>, <span class="pmbm">Czerkies M</span>, <span class="pmbm">Jaruszewicz-Błońska J</span>, <span class="pmbm">Prus W</span>, <span class="pmbm">Kosiuk I</span>, <span class="pmbm">Kochańczyk M</span>, <span class="pmbm">Lipniacki T</span>. '''Non-self RNA rewires IFNβ signaling: A mathematical model of the innate immune response''' (accepted at ''Sci Signal'') [https://doi.org/10.1101/2022.01.30.478391 bioRxiv]<br/>[[File:article_non-self_1.png|300px|link=https://doi.org/10.1101/2022.01.30.478391]]<br/>[[File:article_non-self_2.png|300px|link=https://doi.org/10.1101/2022.01.30.478391]]<br/><br/> | ||
====Other==== | ====Other==== | ||
* Lecture: ''SARS-CoV-2 super-spreaders and super-spreading Variant of Concern 202012/01'', University of Warsaw, Faculty of Physics (Tomasz Lipniacki) | * Lecture: ''SARS-CoV-2 super-spreaders and super-spreading Variant of Concern 202012/01'', University of Warsaw, Faculty of Physics (Tomasz Lipniacki) | ||
| Line 75: | Line 79: | ||
=== | ===Dissemination=== | ||
==== | ====Press==== | ||
* Gazeta Wyborcza, interview, March 2020 ([https://wyborcza.pl/7,75400,25824141,matematyczny-model-rozwoju-epidemii-prof-lipniacki-ograniczenia.html Polish] | [https://wyborcza.pl/7,173236,25831548,poland-follows-down-the-italian-path-prof-lipniacki-author.html English]) | * Gazeta Wyborcza, interview, March 2020 ([https://wyborcza.pl/7,75400,25824141,matematyczny-model-rozwoju-epidemii-prof-lipniacki-ograniczenia.html Polish] | [https://wyborcza.pl/7,173236,25831548,poland-follows-down-the-italian-path-prof-lipniacki-author.html English]) | ||
* Gazeta Wyborcza, interview, June 2020 ([https://wyborcza.pl/7,75400,25920600,prof-lipniacki-musimy-wiedziec-kto-i-jak-sie-zakaza-aby.html Polish]) | * Gazeta Wyborcza, interview, June 2020 ([https://wyborcza.pl/7,75400,25920600,prof-lipniacki-musimy-wiedziec-kto-i-jak-sie-zakaza-aby.html Polish]) | ||
| Line 90: | Line 94: | ||
* Polsat News, debate on 4th wave of COVID-19, August 2021 ([https://www.youtube.com/watch?v=rFn_J6nDjo0&t=1554s Polish]) | * Polsat News, debate on 4th wave of COVID-19, August 2021 ([https://www.youtube.com/watch?v=rFn_J6nDjo0&t=1554s Polish]) | ||
* Radio TOK FM, three separate conversations ([https://audycje.tokfm.pl/gosc/13792,prof-Tomasz-Lipniacki Polish]) | * Radio TOK FM, three separate conversations ([https://audycje.tokfm.pl/gosc/13792,prof-Tomasz-Lipniacki Polish]) | ||
====Public lectures==== | |||
* lecture in German Historical Institute Warsaw ([https://www.dhi.waw.pl/pl/wydarzenia/wyklady/detale/prof-tomasz-lipniacki-virale-invasionen-in-geweben-und-gesellschaften]) | |||
| Line 109: | Line 116: | ||
<br/> | <br/> | ||
[[File:Logo_Norway_Grants.png|88px|left|link=|alt=]] | [[File:Logo_Norway_Grants.png|88px|left|link=|alt=]] | ||
<span style="font-size:x-large;">''Oddziaływanie komórek NK z populacją zainfekowanych komórek rakowych: eksperymenty w urządzeniach mikroprzepływowych i modelowanie matematyczne na poziomie pojedynczych komórek''</span> | <span style="font-size:x-large;">''Oddziaływanie komórek NK z populacją zainfekowanych komórek rakowych: eksperymenty w urządzeniach mikroprzepływowych i modelowanie matematyczne na poziomie pojedynczych komórek''</span> | ||
| Line 148: | Line 155: | ||
===Wyniki=== | ===Wyniki=== | ||
====Publikacje==== | ====Publikacje==== | ||
* <span class="pmbm">Grabowski F</span>, <span class="pmbm">Kochańczyk M</span>, <span class="pmbm">Korwek Z</span>, <span class="pmbm">Czerkies M</span>, <span class="pmbm">Prus W</span>, <span class="pmbm">Lipniacki T</span>. '''Antagonism between viral infection and innate immunity at the single-cell level''', ''PLOS Pathog'' <u>19</u>(9):e1011597 (2023) [https://pubmed.ncbi.nlm.nih.gov/37669278 PubMed] [https://doi.org/10.1371/journal.ppat.1011597 CrossRef] | [https://doi.org/10.1101/2022.11.18.517110 bioRxiv] [https://github.com/grfrederic/visavis Code] [https://doi.org/10.5281/zenodo.7428925 Data]<br/><br/> | |||
* <span class="pmbm">Kurniawan T</span>, <span class="nopmbm">Sahebdivani M</span>, <span class="pmbm">Zaremba D</span>, <span class="pmbm">Blonski S</span>, <span class="nopmbm">Garstecki P</span>, <span class="nopmbm">van Steijn V</span>, <span class="pmbm">Korczyk PM</span>. '''Formation of droplets in microfluidic cross-junctions at small capillary numbers: Breakdown of the classical squeezing regime''', ''Chem Eng J'' <u>474</u>:145601 (2023) [https://doi.org/10.1016/j.cej.2023.145601 CrossRef]<br/><br/> | |||
* <span class="pmbm">Grabowski F</span>, <span class="pmbm">Nałęcz-Jawecki P</span>, <span class="pmbm">Lipniacki T</span>. '''Predictive power of non-identifiable models''', ''Sci Rep'' <u>13</u>:11143 (2023) [https://doi.org/10.1038/s41598-023-37939-8 CrossRef] [https://doi.org/10.1101/2023.04.07.536025 bioRxiv] [https://github.com/grfrederic/identifiability Code]<br/><br/> | |||
* <span class="nopmbm">Sønstevold L</span>, <span class="pmbm">Czerkies M</span>, <span class="nopmbm">Escobedo-Cousin E</span>, <span class="pmbm">Blonski S</span>, <span class="nopmbm">Vereshchagina E</span>. '''Application of polymethylpentene, an oxygen permeable thermoplastic, for long-term on-a-chip cell culture and organ-on-a-chip devices''', ''Micromachines'' <u>14</u>(3):532 (2023) [https://dx.doi.org/10.3390/mi14030532 CrossRef]<br/><br/> | |||
* <span class="author">Czerkies M</span>, <span class="author">Kochańczyk M</span>, <span class="author">Korwek Z</span>, <span class="author">Prus W</span>, <span class="author">Lipniacki T</span>. '''Respiratory syncytial virus protects bystander cells against influenza A virus infection by triggering secretion of type I and type III interferons''', ''J Virol'' (2022) <u>96</u>:22 e01341-22 [https://doi.org/10.1128/jvi.01341-22 CrossRef] [https://doi.org/10.1101/2021.10.11.463877 bioRxiv]<br/>[[File:article_RSV_IAV_crossprotection.png|450px|link=https://doi.org/10.1128/jvi.01341-22]]<br/><br/> | * <span class="author">Czerkies M</span>, <span class="author">Kochańczyk M</span>, <span class="author">Korwek Z</span>, <span class="author">Prus W</span>, <span class="author">Lipniacki T</span>. '''Respiratory syncytial virus protects bystander cells against influenza A virus infection by triggering secretion of type I and type III interferons''', ''J Virol'' (2022) <u>96</u>:22 e01341-22 [https://doi.org/10.1128/jvi.01341-22 CrossRef] [https://doi.org/10.1101/2021.10.11.463877 bioRxiv]<br/>[[File:article_RSV_IAV_crossprotection.png|450px|link=https://doi.org/10.1128/jvi.01341-22]]<br/><br/> | ||
* <span class="pmbm">Grabowski F</span>, <span class="nopmbm">Preibisch G</span>, <span class="nopmbm">Giziński S</span>, <span class="pmbm">Kochańczyk M</span>, <span class="pmbm">Lipniacki T</span>. '''SARS-CoV-2 Variant of Concern 202012/01 has about twofold replicative advantage and acquires concerning mutations''', ''Viruses'' <u>13</u>(3):392 (2021) [https://doi.org/10.3390/v13030392 CrossRef] [https://doi.org/10.1101/2020.12.28.20248906 medRxiv] | [https://www.mdpi.com/1999-4915/13/3/392/pdf PDF]<br/>[[File:article_alpha_1.png|x175px|link=https://doi.org/10.3390/v13030392]] [[File:article_alpha_2.png|x175px|link=https://doi.org/10.3390/v13030392]]<br/><br/> | * <span class="pmbm">Grabowski F</span>, <span class="nopmbm">Preibisch G</span>, <span class="nopmbm">Giziński S</span>, <span class="pmbm">Kochańczyk M</span>, <span class="pmbm">Lipniacki T</span>. '''SARS-CoV-2 Variant of Concern 202012/01 has about twofold replicative advantage and acquires concerning mutations''', ''Viruses'' <u>13</u>(3):392 (2021) [https://doi.org/10.3390/v13030392 CrossRef] [https://doi.org/10.1101/2020.12.28.20248906 medRxiv] | [https://www.mdpi.com/1999-4915/13/3/392/pdf PDF]<br/>[[File:article_alpha_1.png|x175px|link=https://doi.org/10.3390/v13030392]] [[File:article_alpha_2.png|x175px|link=https://doi.org/10.3390/v13030392]]<br/><br/> | ||
* <span class="pmbm">Grabowski F</span>, <span class="pmbm">Kochańczyk M</span>, <span class="pmbm">Lipniacki T</span>. '''The spread of SARS-CoV-2 variant Omicron with the doubling time of 2.0–3.3 days can be explained by immune evasion''', ''Viruses'' <u>14</u>(2):294 (2022) [https://doi.org/10.3390/v14020294 CrossRef] [https://dx.doi.org/10.1101/2021.12.08.21267494 medRxiv] | [https://www.mdpi.com/1999-4915/14/2/294/pdf PDF] [https://susy.mdpi.com/user/review/displayFile/23340271/tDGR6nk0?file=author-coverletter&report=16624478 Korespondencja]<br/>[[File:article_omicron_1.png|x175px|link=https://doi.org/10.3390/v14020294]]<br/><br/> | * <span class="pmbm">Grabowski F</span>, <span class="pmbm">Kochańczyk M</span>, <span class="pmbm">Lipniacki T</span>. '''The spread of SARS-CoV-2 variant Omicron with the doubling time of 2.0–3.3 days can be explained by immune evasion''', ''Viruses'' <u>14</u>(2):294 (2022) [https://doi.org/10.3390/v14020294 CrossRef] [https://dx.doi.org/10.1101/2021.12.08.21267494 medRxiv] | [https://www.mdpi.com/1999-4915/14/2/294/pdf PDF] [https://susy.mdpi.com/user/review/displayFile/23340271/tDGR6nk0?file=author-coverletter&report=16624478 Korespondencja]<br/>[[File:article_omicron_1.png|x175px|link=https://doi.org/10.3390/v14020294]]<br/><br/> | ||
* <span class="nopmbm">Mines RC</span>, <span class="pmbm">Lipniacki T</span>, <span class="nopmbm">Shen X</span>. '''Slow nucleosome dynamics set the transcriptional speed limit and induce RNA polymerase II traffic jams and bursts''', ''PLOS Comput Biol'' <u>18</u>(2):e1009811 (2022) [https://doi.org/10.1371/journal.pcbi.1009811 CrossRef]<br/>[[File:article_nucleodyn_1.png|175px|link=https://doi.org/10.1371/journal.pcbi.1009811]]<br/><br/> | * <span class="nopmbm">Mines RC</span>, <span class="pmbm">Lipniacki T</span>, <span class="nopmbm">Shen X</span>. '''Slow nucleosome dynamics set the transcriptional speed limit and induce RNA polymerase II traffic jams and bursts''', ''PLOS Comput Biol'' <u>18</u>(2):e1009811 (2022) [https://doi.org/10.1371/journal.pcbi.1009811 CrossRef]<br/>[[File:article_nucleodyn_1.png|175px|link=https://doi.org/10.1371/journal.pcbi.1009811]]<br/><br/> | ||
* <span class="pmbm">Korwek Z</span>, <span class="pmbm">Czerkies M</span>, <span class="pmbm">Jaruszewicz-Błońska J</span>, <span class="pmbm">Prus W</span>, <span class="pmbm">Kosiuk I</span>, <span class="pmbm">Kochańczyk M</span>, <span class="pmbm">Lipniacki T</span>. '''Non-self RNA rewires IFNβ signaling: A mathematical model of the innate immune response''' ( | * <span class="pmbm">Korwek Z</span>, <span class="pmbm">Czerkies M</span>, <span class="pmbm">Jaruszewicz-Błońska J</span>, <span class="pmbm">Prus W</span>, <span class="pmbm">Kosiuk I</span>, <span class="pmbm">Kochańczyk M</span>, <span class="pmbm">Lipniacki T</span>. '''Non-self RNA rewires IFNβ signaling: A mathematical model of the innate immune response''' (przyjęte w ''Sci Signal'') [https://doi.org/10.1101/2022.01.30.478391 bioRxiv]<br/>[[File:article_non-self_1.png|300px]]<br/>[[File:article_non-self_2.png|300px|link=https://doi.org/10.1101/2022.01.30.478391]]<br/><br/> | ||
====Inne==== | ====Inne==== | ||
* Wykład: ''SARS-CoV-2 super-spreaders and super-spreading Variant of Concern 202012/01'', Uniwersytet Warszawski, Wydział Fizyki (Tomasz Lipniacki) | * Wykład: ''SARS-CoV-2 super-spreaders and super-spreading Variant of Concern 202012/01'', Uniwersytet Warszawski, Wydział Fizyki (Tomasz Lipniacki) | ||
| Line 165: | Line 176: | ||
Wraz z naszymi współpracownikami z SINTEF AS badamy działanie termoplastycznego materiału PMP w urządzeniach mikroprzepływowych do organ-on-a-chip i długoterminowej hodowli komórek na chipach. Ocena obejmuje badanie przylegania i proliferacji komórek, przydatność materiału do mikroskopii konfokalnej, zgodność z powszechnie stosowanymi substancjami chemicznymi, wytwarzanie urządzeń i, co najważniejsze, ocenę przepuszczalności gazu dla dostarczania tlenu podczas 4 dni hodowli komórek. | Wraz z naszymi współpracownikami z SINTEF AS badamy działanie termoplastycznego materiału PMP w urządzeniach mikroprzepływowych do organ-on-a-chip i długoterminowej hodowli komórek na chipach. Ocena obejmuje badanie przylegania i proliferacji komórek, przydatność materiału do mikroskopii konfokalnej, zgodność z powszechnie stosowanymi substancjami chemicznymi, wytwarzanie urządzeń i, co najważniejsze, ocenę przepuszczalności gazu dla dostarczania tlenu podczas 4 dni hodowli komórek. | ||
[[ | [[Image:PMP_device_1.png|x200px]] | ||
[[File:PMP_device_2.png|x200px]] | [[File:PMP_device_2.png|x200px]] | ||
<br/> | <br/> | ||
| Line 172: | Line 183: | ||
=== | ===Popularyzacja=== | ||
==== | ====Prasa==== | ||
* Gazeta Wyborcza, wywiad, marzec 2020 ([https://wyborcza.pl/7,75400,25824141,matematyczny-model-rozwoju-epidemii-prof-lipniacki-ograniczenia.html po polsku], [https://wyborcza.pl/7,173236,25831548,poland-follows-down-the-italian-path-prof-lipniacki-author.html po angielsku]) | * Gazeta Wyborcza, wywiad, marzec 2020 ([https://wyborcza.pl/7,75400,25824141,matematyczny-model-rozwoju-epidemii-prof-lipniacki-ograniczenia.html po polsku], [https://wyborcza.pl/7,173236,25831548,poland-follows-down-the-italian-path-prof-lipniacki-author.html po angielsku]) | ||
* Gazeta Wyborcza, wywiad, czerwiec 2020 ([https://wyborcza.pl/7,75400,25920600,prof-lipniacki-musimy-wiedziec-kto-i-jak-sie-zakaza-aby.html po polsku]) | * Gazeta Wyborcza, wywiad, czerwiec 2020 ([https://wyborcza.pl/7,75400,25920600,prof-lipniacki-musimy-wiedziec-kto-i-jak-sie-zakaza-aby.html po polsku]) | ||
| Line 181: | Line 192: | ||
* Gazeta Wyborcza, wywiad, lipiec 2021 ([https://wyborcza.pl/7,75400,26665472,prof-lipniacki-sars-cov-2-po-raz-kolejny-znaczaco-zmutowal.html po polsku]) | * Gazeta Wyborcza, wywiad, lipiec 2021 ([https://wyborcza.pl/7,75400,26665472,prof-lipniacki-sars-cov-2-po-raz-kolejny-znaczaco-zmutowal.html po polsku]) | ||
====Telewizja | ====Radio i Telewizja==== | ||
* Polsat News, debata o rozwoju pandemii i roli szczepień, listopad 2020 ([https://www.youtube.com/watch?v=KQIvgg9Oopo&t=391 po polsku]) | * Polsat News, debata o rozwoju pandemii i roli szczepień, listopad 2020 ([https://www.youtube.com/watch?v=KQIvgg9Oopo&t=391 po polsku]) | ||
* Polsat News, debata o restrykcjach sylwestrowych, grudzień 2020 ([https://www.youtube.com/watch?v=EvZq4QO9CVw&t=75s po polsku]) | * Polsat News, debata o restrykcjach sylwestrowych, grudzień 2020 ([https://www.youtube.com/watch?v=EvZq4QO9CVw&t=75s po polsku]) | ||
| Line 187: | Line 198: | ||
* Polsat News, debata o czwartej fali COVID-19, sierpień 2021 ([https://www.youtube.com/watch?v=rFn_J6nDjo0&t=1554s po polsku]) | * Polsat News, debata o czwartej fali COVID-19, sierpień 2021 ([https://www.youtube.com/watch?v=rFn_J6nDjo0&t=1554s po polsku]) | ||
* Radio TOK FM, trzy rozmowy ([https://audycje.tokfm.pl/gosc/13792,prof-Tomasz-Lipniacki po polsku]) | * Radio TOK FM, trzy rozmowy ([https://audycje.tokfm.pl/gosc/13792,prof-Tomasz-Lipniacki po polsku]) | ||
===Inne=== | |||
* wykład w Niemieckim Instytucie Historycznym w Warszawie ([https://www.dhi.waw.pl/pl/wydarzenia/wyklady/detale/prof-tomasz-lipniacki-virale-invasionen-in-geweben-und-gesellschaften]) | |||
Latest revision as of 20:49, 22 October 2023

Interaction of natural killer cells with infected cancer cell population: microfluidics-based experiments and single-cell-level mathematical modeling
Concepts and objectives
The aim of the project is to analyze interactions of natural killer (NK) cells with an infected epithelial cell population at the single-cell resolution. We investigate two respiratory epithelial cell lines: cancerous and non-cancerous, and two respiratory viruses: respiratory syncytial virus (RSV) and influenza A virus (IAV). Our main objectives are as follows:
- Elucidation of the impact of the NK cell-induced cell death on the dynamics of propagation and eradication of RSV and IAV infections.
- Determination of the potential of NK cells to induce immunogenic cell death in infected cells, that can potentially activate adaptive immune response.
- Development of a droplet-based microfluidic system to perform experiments on co-cultures of epithelial and NK cells (living in droplets), that will enable data acquisition at the single-cell resolution.
- Development of a mathematical model capturing interactions of NK cells with an infected cell population at the single-cell resolution.
Team

IPPT PAN:
- Prof. Tomasz Lipniacki, PhD (head)
- Sławomir Błoński, PhD
- Maciej Czerkies, PhD
- Frederic Grabowski, BSc
- Joanna Jaruszewicz-Błońska, PhD
- Marek Kochańczyk, PhD
- Piotr Korczyk, PhD
- Zbigniew Korwek, PhD
- Paulina Koza, PhD
- Barbara Kupikowska-Stobba, PhD
- Felipe Nieves Marques Porto, MSc
- Wiktor Prus, PhD
- Damian Zaremba, MSc
SINTEF AS (core team):
- Elizaveta Vereshchagina, PhD (head)
- Linda Sønstevold, PhD
- Michal Marek Mielnik, PhD
- Geir Uri Jensen, PhD
- Shruti Jain, MSc
- Enrique Escobedo-Cousin, PhD
Research
Scientific publications
- Grabowski F, Kochańczyk M, Korwek Z, Czerkies M, Prus W, Lipniacki T. Antagonism between viral infection and innate immunity at the single-cell level, PLOS Pathog 19(9):e1011597 (2023) PubMed CrossRef | bioRxiv Code Data
- Kurniawan T, Sahebdivani M, Zaremba D, Blonski S, Garstecki P, van Steijn V, Korczyk PM. Formation of droplets in microfluidic cross-junctions at small capillary numbers: Breakdown of the classical squeezing regime, Chem Eng J 474:145601 (2023) CrossRef
- Grabowski F, Nałęcz-Jawecki P, Lipniacki T. Predictive power of non-identifiable models, Sci Rep 13:11143 (2023) CrossRef bioRxiv Code
- Sønstevold L, Czerkies M, Escobedo-Cousin E, Blonski S, Vereshchagina E. Application of polymethylpentene, an oxygen permeable thermoplastic, for long-term on-a-chip cell culture and organ-on-a-chip devices, Micromachines 14(3):532 (2023) CrossRef
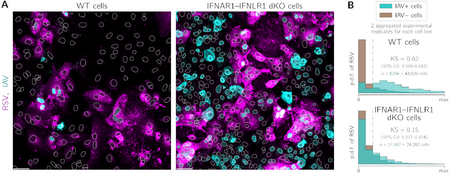
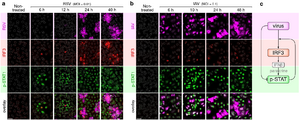
- , , , , . Respiratory syncytial virus protects bystander cells against influenza A virus infection by triggering secretion of type I and type III interferons, J Virol (2022) 96:22 e01341-22 CrossRef bioRxiv
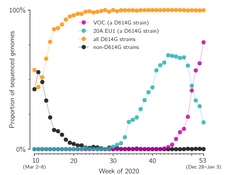
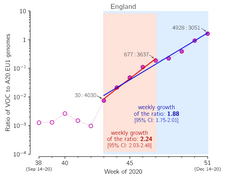
- Grabowski F, Preibisch G, Giziński S, Kochańczyk M, Lipniacki T. SARS-CoV-2 Variant of Concern 202012/01 has about twofold replicative advantage and acquires concerning mutations, Viruses 13(3):392 (2021) CrossRef medRxiv | PDF
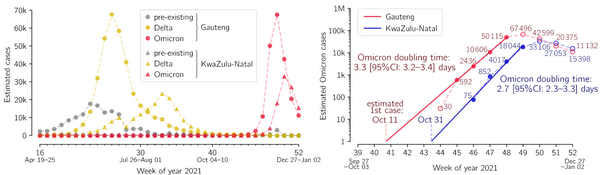
- Grabowski F, Kochańczyk M, Lipniacki T. The spread of SARS-CoV-2 variant Omicron with the doubling time of 2.0–3.3 days can be explained by immune evasion, Viruses 14(2):294 (2022) CrossRef medRxiv | PDF Correspondence
- Mines RC, Lipniacki T, Shen X. Slow nucleosome dynamics set the transcriptional speed limit and induce RNA polymerase II traffic jams and bursts, PLOS Comput Biol 18(2):e1009811 (2022) CrossRef
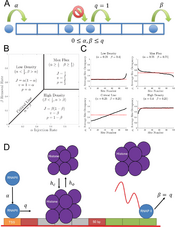
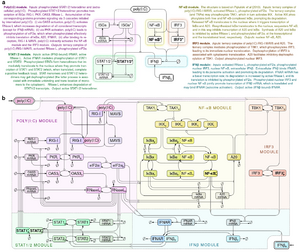
- Korwek Z, Czerkies M, Jaruszewicz-Błońska J, Prus W, Kosiuk I, Kochańczyk M, Lipniacki T. Non-self RNA rewires IFNβ signaling: A mathematical model of the innate immune response (accepted at Sci Signal) bioRxiv
Other
- Lecture: SARS-CoV-2 super-spreaders and super-spreading Variant of Concern 202012/01, University of Warsaw, Faculty of Physics (Tomasz Lipniacki)
- Lecture: SARS‐CoV‐2 Variant of Concern 202012/01 has about twofold replicative advantage and acquires concerning mutations, University of Warsaw, Faculty of Mathematics, Informatics and Mechanics, BOB seminar (Frederic Grabowski)
- Keynote lecture: Mathematical modeling of innate immune response to viral infection, Joint KMM-VIN / ViCEM / ESB cross-disciplinary workshop, Vienna, Austria 2022 (Tomasz Lipniacki) Conference website
- Talk: Dissecting innate immunity responses to viral infection at the single-cell level, Inverse Problems Meeting (virtual conference) Video
- Talk: Non-self RNA rewires IFNβ signaling: A model of the innate immune response, q-Bio 2022, Colorado State University (Tomasz Lipniacki) Abstract Slides
Technology
With our collaborators from SINTEF AS we investigate the performance of the thermoplastic material PMP in microfluidic devices for organ-on-a-chip and long-term cell cultivation on chip applications. The assessment includes investigating cell adherence and proliferation, material suitability for confocal microscopy, compatibility with common chemicals, device fabrication, and most importantly assessment of gas permeability for oxygen supply during 4 days of cell culturing.


Device used in experiments (article under preparation).
Dissemination
Press
- Gazeta Wyborcza, interview, March 2020 (Polish | English)
- Gazeta Wyborcza, interview, June 2020 (Polish)
- Newsweek Polska, interview, October 2020 (Polish)
- Gazeta Wyborcza, article, October 2020 (Polish)
- Gazeta Wyborcza, article, November 2020 (Polish)
- Gazeta Wyborcza, interview, July 2021 (Polish)
Broadcast
- Polsat News, debate on pandemic development and role of vaccinations, November 2020 (Polish)
- Polsat News, debate on New Year restrictions, December 2020 (Polish)
- TVN24, debate on lockdown strategy, April 2021 (Polish)
- Polsat News, debate on 4th wave of COVID-19, August 2021 (Polish)
- Radio TOK FM, three separate conversations (Polish)
Public lectures
- lecture in German Historical Institute Warsaw ([1])
Contact
Lab head – Prof. Tomasz Lipniacki.
Funding
This research is funded by the Norwegian Financial Mechanism GRIEG-1
(grant 2019/34/H/NZ6/00699, operated by the National Science Centre, Poland).

Oddziaływanie komórek NK z populacją zainfekowanych komórek rakowych: eksperymenty w urządzeniach mikroprzepływowych i modelowanie matematyczne na poziomie pojedynczych komórek
Cele projektu
Celem projektu jest analiza oddziaływań komórek “Naturalnych Zabójców” (ang. Natural Killer, NK) z zakażonymi rakowymi komórkami nabłonka płuc na poziomie pojedynczych komórek. Badamy dwie linie komórek nabłonka oddechowego, z których jedna ma pochodzenie nowotworowe oraz dwa wirusy: syncytialny wirus nabłonka oddechowego (RSV) oraz wirus grypy typu A (IAV). Nasze główne zadania badawcze są następujące:
- wyjaśnienie wpływu cytotoksyczności komórek NK wobec zakażonych komórek na dynamikę rozprzestrzeniania się oraz eliminacji infekcji wirusami RSV i IAV;
- ustalenie potencjału komórek NK do indukowania w zakażonych komórkach tzw. immunogennej śmierci komórkowej zdolnej do aktywacji mechanizmów nabytej obrony odpornościowej;
- rozwój systemu mikrofluidyki kropelkowej przeznaczonej prowadzenia doświadczeń na kokulturach komórek nabłonkowych i komórek NK, który pozwoli na zbieranie danych na poziomie poszczególnych komórek;
- rozwój modelu matematycznego opisującego interakcje komórek NK z zainfekowaną populacją komórek nabłonkowych, na poziomie pojedynczych komórek.
Zespół

IPPT PAN:
- prof. Tomasz Lipniacki (kierownik projektu)
- dr Wiktor Prus
- dr Maciej Czerkies
- dr Zbigniew Korwek
- dr Joanna Jaruszewicz-Błońska
- dr Marek Kochańczyk
- dr Paulina Koza
- mgr Damian Zaremba
- mgr Felipe Nieves Marques Porto
- lic. Frederic Grabowski
SINTEF AS (core team):
- dr Elizaveta Vereshchagina (koordynator strony norweskiej)
- dr Linda Sønstevold
- dr Michal Marek Mielnik
- dr Geir Uri Jensen
- mgr Shruti Jain
- dr Enrique Escobedo-Cousin
Wyniki
Publikacje
- Grabowski F, Kochańczyk M, Korwek Z, Czerkies M, Prus W, Lipniacki T. Antagonism between viral infection and innate immunity at the single-cell level, PLOS Pathog 19(9):e1011597 (2023) PubMed CrossRef | bioRxiv Code Data
- Kurniawan T, Sahebdivani M, Zaremba D, Blonski S, Garstecki P, van Steijn V, Korczyk PM. Formation of droplets in microfluidic cross-junctions at small capillary numbers: Breakdown of the classical squeezing regime, Chem Eng J 474:145601 (2023) CrossRef
- Grabowski F, Nałęcz-Jawecki P, Lipniacki T. Predictive power of non-identifiable models, Sci Rep 13:11143 (2023) CrossRef bioRxiv Code
- Sønstevold L, Czerkies M, Escobedo-Cousin E, Blonski S, Vereshchagina E. Application of polymethylpentene, an oxygen permeable thermoplastic, for long-term on-a-chip cell culture and organ-on-a-chip devices, Micromachines 14(3):532 (2023) CrossRef
- , , , , . Respiratory syncytial virus protects bystander cells against influenza A virus infection by triggering secretion of type I and type III interferons, J Virol (2022) 96:22 e01341-22 CrossRef bioRxiv

- Grabowski F, Preibisch G, Giziński S, Kochańczyk M, Lipniacki T. SARS-CoV-2 Variant of Concern 202012/01 has about twofold replicative advantage and acquires concerning mutations, Viruses 13(3):392 (2021) CrossRef medRxiv | PDF


- Grabowski F, Kochańczyk M, Lipniacki T. The spread of SARS-CoV-2 variant Omicron with the doubling time of 2.0–3.3 days can be explained by immune evasion, Viruses 14(2):294 (2022) CrossRef medRxiv | PDF Korespondencja

- Mines RC, Lipniacki T, Shen X. Slow nucleosome dynamics set the transcriptional speed limit and induce RNA polymerase II traffic jams and bursts, PLOS Comput Biol 18(2):e1009811 (2022) CrossRef

- Korwek Z, Czerkies M, Jaruszewicz-Błońska J, Prus W, Kosiuk I, Kochańczyk M, Lipniacki T. Non-self RNA rewires IFNβ signaling: A mathematical model of the innate immune response (przyjęte w Sci Signal) bioRxiv


Inne
- Wykład: SARS-CoV-2 super-spreaders and super-spreading Variant of Concern 202012/01, Uniwersytet Warszawski, Wydział Fizyki (Tomasz Lipniacki)
- Wykład: SARS‐CoV‐2 Variant of Concern 202012/01 has about twofold replicative advantage and acquires concerning mutations, Uniwersytet Warszawski, Wydział Matematyki, Informatyki i Mechaniki, seminarium BOB (Frederic Grabowski)
- Wykład zaproszony: Mathematical modeling of innate immune response to viral infection, Joint KMM-VIN / ViCEM / ESB cross-disciplinary workshop, Vienna, Austria 2022 (Tomasz Lipniacki) strona konferencji
- Wystąpienie: Dissecting innate immunity responses to viral infection at the single-cell level, Inverse Problems Meeting (virtual conference) dostępne online
- Wystąpienie: Non-self RNA rewires IFNβ signaling: A model of the innate immune response, q-Bio 2022, Colorado State University (Tomasz Lipniacki) podsumowanie Slajdy
Technologia
Wraz z naszymi współpracownikami z SINTEF AS badamy działanie termoplastycznego materiału PMP w urządzeniach mikroprzepływowych do organ-on-a-chip i długoterminowej hodowli komórek na chipach. Ocena obejmuje badanie przylegania i proliferacji komórek, przydatność materiału do mikroskopii konfokalnej, zgodność z powszechnie stosowanymi substancjami chemicznymi, wytwarzanie urządzeń i, co najważniejsze, ocenę przepuszczalności gazu dla dostarczania tlenu podczas 4 dni hodowli komórek.


Urządzenie używane w eksperymentach (publikacja w przygotowaniu).
Popularyzacja
Prasa
- Gazeta Wyborcza, wywiad, marzec 2020 (po polsku, po angielsku)
- Gazeta Wyborcza, wywiad, czerwiec 2020 (po polsku)
- Newsweek Polska, wywiad, październik 2020 (po polsku)
- Gazeta Wyborcza, artykuł, październik 2020 (po polsku)
- Gazeta Wyborcza, artykuł, listopad 2020 (po polsku)
- Gazeta Wyborcza, wywiad, lipiec 2021 (po polsku)
Radio i Telewizja
- Polsat News, debata o rozwoju pandemii i roli szczepień, listopad 2020 (po polsku)
- Polsat News, debata o restrykcjach sylwestrowych, grudzień 2020 (po polsku)
- TVN24, debata o strategii lockdownów, kwiecień 2021 (po polsku)
- Polsat News, debata o czwartej fali COVID-19, sierpień 2021 (po polsku)
- Radio TOK FM, trzy rozmowy (po polsku)
Inne
- wykład w Niemieckim Instytucie Historycznym w Warszawie ([2])
Kontakt
Kierownik Pracowni – Prof. Tomasz Lipniacki.
Finansowanie
Badania są finansowane z Norweskiego Mechanizmu Finansowego GRIEG-1
(grant 2019/34/H/NZ6/00699, którego operatorem jest Narodowe Centrum Nauki).
